|
PLearn 0.1
|
|
PLearn 0.1
|
Implements an online natural gradient, based on keeping an estimate of the gradients' covariance C through its main eigen vectors and values which are updated through those of the gram matrix. More...
#include <OnlineGramNaturalGradientOptimizer.h>
Public Member Functions | |
| OnlineGramNaturalGradientOptimizer () | |
| void | gramEigenNaturalGradient () |
| virtual bool | optimizeN (VecStatsCollector &stats_coll) |
| Main optimization method, to be defined in subclasses. | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual OnlineGramNaturalGradientOptimizer * | deepCopy (CopiesMap &copies) const |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Does the necessary operations to transform a shallow copy (this) into a deep copy by deep-copying all the members that need to be. | |
| virtual void | build () |
| Post-constructor. | |
Static Public Member Functions | |
| static string | _classname_ () |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| real | learning_rate |
| real | gamma |
| real | reg |
| int | opt_batch_size |
| int | n_eigen |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares the class options. | |
Private Types | |
| typedef Optimizer | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
Private Attributes | |
| int | n_optimizeN_calls |
| int | bi |
| int | n_eigen_cur |
| int | n_eigen_old |
| Mat | gradients |
| Vec | mu |
| real | total_variance |
| real | variance_percentage |
| Mat | gram |
| Mat | U |
| Vec | D |
| Mat | cov_eigen_vec |
| Mat | old_cov_eigen_vec |
| Mat | swap |
| Vec | cov_eigen_val |
| Mat | cov_norm_eigen_vec |
| Vec | dot_prod |
| Vec | scaled_dot_prod |
| Vec | naturalg |
| clock_t | t0 |
| clock_t | t1 |
| clock_t | t2 |
| clock_t | t3 |
Implements an online natural gradient, based on keeping an estimate of the gradients' covariance C through its main eigen vectors and values which are updated through those of the gram matrix.
This is n_eigen^2 instead of n_parameter^2.
Definition at line 59 of file OnlineGramNaturalGradientOptimizer.h.
typedef Optimizer PLearn::OnlineGramNaturalGradientOptimizer::inherited [private] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.h.
| PLearn::OnlineGramNaturalGradientOptimizer::OnlineGramNaturalGradientOptimizer | ( | ) |
Definition at line 63 of file OnlineGramNaturalGradientOptimizer.cc.
:
learning_rate(0.01),
gamma(1.0),
reg(1e-6),
opt_batch_size(1),
n_eigen(6)
{}
| string PLearn::OnlineGramNaturalGradientOptimizer::_classname_ | ( | ) | [static] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| OptionList & PLearn::OnlineGramNaturalGradientOptimizer::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| RemoteMethodMap & PLearn::OnlineGramNaturalGradientOptimizer::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| Object * PLearn::OnlineGramNaturalGradientOptimizer::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| StaticInitializer OnlineGramNaturalGradientOptimizer::_static_initializer_ & PLearn::OnlineGramNaturalGradientOptimizer::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| virtual void PLearn::OnlineGramNaturalGradientOptimizer::build | ( | ) | [inline, virtual] |
Post-constructor.
The normal implementation should call simply inherited::build(), then this class's build_(). This method should be callable again at later times, after modifying some option fields to change the "architecture" of the object.
Reimplemented from PLearn::Optimizer.
Definition at line 89 of file OnlineGramNaturalGradientOptimizer.h.
{
inherited::build();
build_();
}
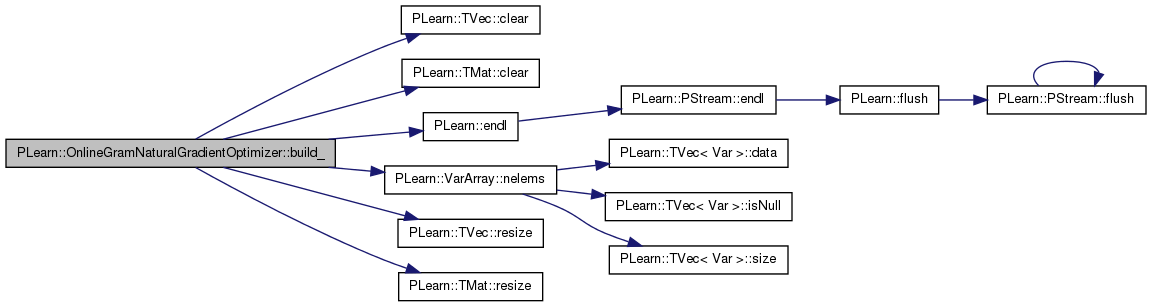
| void PLearn::OnlineGramNaturalGradientOptimizer::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::Optimizer.
Definition at line 116 of file OnlineGramNaturalGradientOptimizer.cc.
References PLearn::TVec< T >::clear(), PLearn::TMat< T >::clear(), PLearn::endl(), gradients, mu, n, n_eigen_cur, n_eigen_old, n_optimizeN_calls, naturalg, PLearn::VarArray::nelems(), opt_batch_size, PLearn::Optimizer::params, PLearn::TVec< T >::resize(), PLearn::TMat< T >::resize(), total_variance, and variance_percentage.
{
n_optimizeN_calls=0;
n_eigen_cur = 0;
n_eigen_old = 0;
total_variance = 0.0;
variance_percentage = 0.;
int n = params.nelems();
cout << "Number of parameters: " << n << endl;
if (n > 0) {
gradients.resize( opt_batch_size, n );
gradients.clear();
mu.resize(n);
mu.clear();
naturalg.resize(n);
naturalg.clear();
// other variables will have different lengths
// depending on the current number of eigen vectors
}
}
| string PLearn::OnlineGramNaturalGradientOptimizer::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
Referenced by optimizeN().

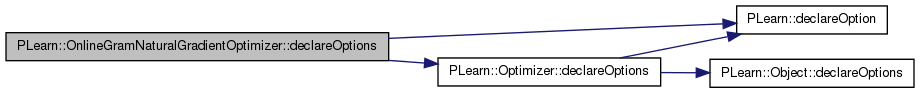
| void PLearn::OnlineGramNaturalGradientOptimizer::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares the class options.
Reimplemented from PLearn::Optimizer.
Definition at line 72 of file OnlineGramNaturalGradientOptimizer.cc.
References PLearn::OptionBase::buildoption, PLearn::declareOption(), PLearn::Optimizer::declareOptions(), gamma, learning_rate, n_eigen, opt_batch_size, and reg.
{
declareOption(
ol, "learning_rate", &OnlineGramNaturalGradientOptimizer::learning_rate,
OptionBase::buildoption,
"Learning rate used in the natural gradient descent.\n");
declareOption(
ol, "gamma", &OnlineGramNaturalGradientOptimizer::gamma,
OptionBase::buildoption,
"Discount factor used in the update of the estimate of the gradient covariance.\n");
declareOption(
ol, "reg", &OnlineGramNaturalGradientOptimizer::reg,
OptionBase::buildoption,
"Regularizer used in computing the natural gradient, C^{-1} mu. Added to C^{-1} diagonal.\n");
declareOption(
ol, "opt_batch_size", &OnlineGramNaturalGradientOptimizer::opt_batch_size,
OptionBase::buildoption,
"Size of the optimizer's batches (examples before parameter and gradient covariance updates).\n");
declareOption(
ol, "n_eigen", &OnlineGramNaturalGradientOptimizer::n_eigen,
OptionBase::buildoption,
"The number of eigen vectors to model the gradient covariance matrix\n");
inherited::declareOptions(ol);
}
| static const PPath& PLearn::OnlineGramNaturalGradientOptimizer::declaringFile | ( | ) | [inline, static] |
Reimplemented from PLearn::Optimizer.
Definition at line 85 of file OnlineGramNaturalGradientOptimizer.h.
{
| OnlineGramNaturalGradientOptimizer * PLearn::OnlineGramNaturalGradientOptimizer::deepCopy | ( | CopiesMap & | copies | ) | const [virtual] |
Reimplemented from PLearn::Optimizer.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| OptionList & PLearn::OnlineGramNaturalGradientOptimizer::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| OptionMap & PLearn::OnlineGramNaturalGradientOptimizer::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
| RemoteMethodMap & PLearn::OnlineGramNaturalGradientOptimizer::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 61 of file OnlineGramNaturalGradientOptimizer.cc.
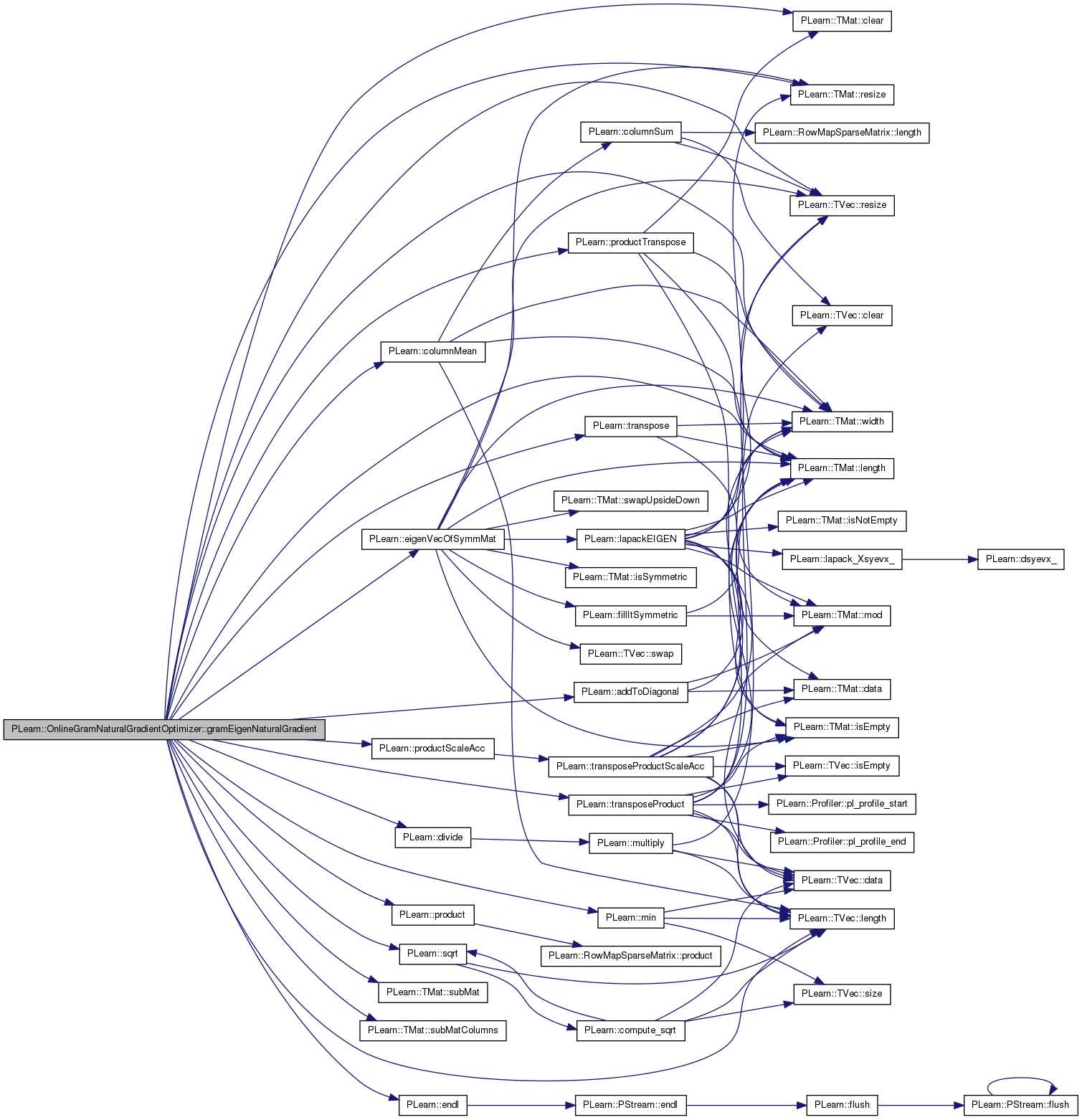
| void PLearn::OnlineGramNaturalGradientOptimizer::gramEigenNaturalGradient | ( | ) |
Definition at line 203 of file OnlineGramNaturalGradientOptimizer.cc.
References PLearn::addToDiagonal(), PLearn::TMat< T >::clear(), PLearn::columnMean(), cov_eigen_val, cov_eigen_vec, cov_norm_eigen_vec, D, PLearn::divide(), dot_prod, PLearn::eigenVecOfSymmMat(), PLearn::endl(), gamma, gradients, gram, i, PLearn::TMat< T >::length(), PLearn::TVec< T >::length(), m, PLearn::min(), mu, n_eigen, n_eigen_cur, n_eigen_old, naturalg, old_cov_eigen_vec, PLWARNING, PLearn::product(), PLearn::productScaleAcc(), PLearn::productTranspose(), reg, PLearn::TMat< T >::resize(), PLearn::TVec< T >::resize(), scaled_dot_prod, PLearn::sqrt(), PLearn::TMat< T >::subMat(), PLearn::TMat< T >::subMatColumns(), swap, PLearn::transpose(), PLearn::transposeProduct(), U, and PLearn::TMat< T >::width().
Referenced by optimizeN().
{
// We don't have any eigen vectors yet
if( n_eigen_cur == 0 ) {
// The number of eigen vectors we will have after incorporating the new data
// (the gram matrix of gradients might have a rank smaller than n_eigen)
n_eigen_cur = min( gradients.length(), n_eigen);
// Compute the total variance - to do this, compute the trace of the covariance matrix
// could also use the trace of the gram matrix since we compute it, ie sum(diag(gram))
/* for( int i=0; i<gradients.length(); i++) {
Vec v = gradients(i);
total_variance += sumsquare(v);
}
total_variance /= gradients.length();*/
// Compute the gram matrix - TODO does this recognize gram is symetric? (and save the computations?)
gram.resize( gradients.length(), gradients.length() );
productTranspose(gram, gradients, gradients);
gram /= gradients.length();
// Extract eigenvectors/eigenvalues - destroys the content of gram, D and U are resized
// gram = U D U' (if we took all values)
eigenVecOfSymmMat(gram, n_eigen_cur, D, U);
// Percentage of the variance we keep is the sum of the kept eigenvalues divided
// by the total variance.
//variance_percentage = sum(D)/total_variance;
// The eigenvectors V of C are deduced from the eigenvectors U of G by the
// formula V = AUD^{-1/2} (D the eigenvalues of G). The nonzero eigenvalues of
// C and D are the same.
// The true eigenvalues are norm_eigen_vec. However, we shall keep in memory
// the eigenvectors of C rescaled by the square root of their associated
// eigenvalues, so that C can be written VV' instead of VDV'. Thus, the "new" V
// is equal to VD^{1/2} = AU.
// We have row vectors so AU = (U'A')'
cov_eigen_vec.resize(n_eigen_cur, gradients.width() );
product( cov_eigen_vec, U, gradients );
cov_eigen_vec /= sqrt( gradients.length() );
cov_eigen_val.resize( D.length() );
cov_eigen_val << D;
ofstream fd_eigval("eigen_vals.txt", ios_base::app);
fd_eigval << cov_eigen_val << endl;
fd_eigval.close();
cov_norm_eigen_vec.resize( n_eigen_cur, gradients.width() );
for( int i=0; i<n_eigen_cur; i++) {
Vec v = cov_norm_eigen_vec(i);
divide( cov_eigen_vec(i), sqrt(D[i]), v );
}
}
// We already have some eigen vectors, so it's an update
else {
// The number of eigen vectors we will have after incorporating the new data
n_eigen_old = cov_eigen_vec.length();
n_eigen_cur = min( cov_eigen_vec.length() + gradients.length(), n_eigen);
// Update the total variance, by computing that of the covariance matrix
// total_variance = gamma*total_variance + (1-gamma)*sum(sum(A.^2))/n_new_vec
/*total_variance *= gamma;
for( int i=0; i<gradients.length(); i++) {
Vec v = gradients(i);
// To reflect the new update
//total_variance += (1.-gamma) * sumsquare(v) / gradients.length();
total_variance += sumsquare(v) / gradients.length();
}*/
// Compute the gram matrix
// To find the equivalence between the covariance matrix and the Gram matrix,
// we need to have the covariance matrix under the form C = UU' + AA'. However,
// what we have is C = gamma UU' + (1-gamma)AA'/n_new_vec. Thus, we will
// rescale U and A using U = sqrt(gamma) U and A = sqrt((1 - gamma)/n_new_vec)
// A. Now, the Gram matrix is of the form [U'U U'A;A'U A'A] using the new U and
// A.
gram.resize( n_eigen_old + gradients.length(), n_eigen_old + gradients.length() );
Mat m = gram.subMat(0, 0, n_eigen_old, n_eigen_old);
m.clear();
addToDiagonal(m, gamma*D);
// Nicolas says "use C_{n+1} = gamma C_n + gg'" so no (1.-gamma)
m = gram.subMat(n_eigen_old, n_eigen_old, gradients.length(), gradients.length());
productTranspose(m, gradients, gradients);
//m *= (1.-gamma) / gradients.length();
m /= gradients.length();
m = gram.subMat(n_eigen_old, 0, gradients.length(), n_eigen_old );
productTranspose(m, gradients, cov_eigen_vec);
//m *= sqrt(gamma*(1.-gamma)/gradients.length());
m *= sqrt(gamma/gradients.length());
Mat m2 = gram.subMat( 0, n_eigen_old, n_eigen_old, gradients.length() );
transpose( m, m2 );
//G = (G + G')/2; % Solving numerical mistakes
//cout << "--" << endl << gram << endl;
// Extract eigenvectors/eigenvalues - destroys the content of gram, D and U are resized
// gram = U D U' (if we took all values)
eigenVecOfSymmMat(gram, n_eigen_cur, D, U);
// Percentage of the variance we keep is the sum of the kept eigenvalues divided
// by the total variance.
//variance_percentage = sum(D)/total_variance;
// The new (rescaled) eigenvectors are of the form [U A]*V where V is the
// eigenvector of G. Rewriting V = [V1;V2], we have [U A]*V = UV1 + AV2.
// for us cov_eigen_vec = U1 eigen_vec + U2 gradients
swap = old_cov_eigen_vec;
old_cov_eigen_vec = cov_eigen_vec;
cov_eigen_vec = swap;
cov_eigen_vec.resize(n_eigen_cur, gradients.width());
product( cov_eigen_vec, U.subMatColumns(0, n_eigen_old), old_cov_eigen_vec );
// C = alpha A.B + beta C
productScaleAcc(cov_eigen_vec, U.subMatColumns(n_eigen_old, gradients.length()), false, gradients, false,
sqrt((1.-gamma)/gradients.length()), sqrt(gamma));
cov_eigen_val.resize( D.length() );
cov_eigen_val << D;
cov_norm_eigen_vec.resize( n_eigen_cur, gradients.width() );
for( int i=0; i<n_eigen_cur; i++) {
Vec v = cov_norm_eigen_vec(i);
divide( cov_eigen_vec(i), sqrt(D[i]), v );
}
}
// ### Determine reg - Should be set automaticaly.
//reg = cov_eigen_val[n_eigen_cur-1];
for( int i=0; i<n_eigen_cur; i++) {
if( cov_eigen_val[i] < reg ) {
PLWARNING("cov_eigen_val[i] < reg. Setting to reg.");
cov_eigen_val[i] = reg;
}
}
// *** Compute C^{-1} mu, where mu is the mean of gradients ***
// Compute mu
columnMean( gradients, mu );
/* cout << "mu " << mu << endl;
cout << "norm(mu) " << norm(mu) << endl;
cout << "cov_eigen_val " << cov_eigen_val << endl;
cout << "cov_eigen_vec " << cov_eigen_vec << endl;
cout << "cov_norm_eigen_vec " << cov_norm_eigen_vec << endl;*/
// Compute the dot product with the eigenvectors
dot_prod.resize(n_eigen_cur);
product( dot_prod, cov_norm_eigen_vec, mu);
// cout << "dot_prod " << dot_prod << endl;
// Rescale according to the eigenvectors. Since the regularization constant will
// be added to all the eigenvalues (and not only the ones we didn't keep), we
// have to remove it from the ones we kept.
scaled_dot_prod.resize(n_eigen_cur);
divide( dot_prod, cov_eigen_val, scaled_dot_prod);
scaled_dot_prod -= dot_prod/reg;
transposeProduct(naturalg, cov_norm_eigen_vec, scaled_dot_prod);
naturalg += mu / reg;
}

| void PLearn::OnlineGramNaturalGradientOptimizer::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Does the necessary operations to transform a shallow copy (this) into a deep copy by deep-copying all the members that need to be.
This needs to be overridden by every class that adds "complex" data members to the class, such as Vec, Mat, PP<Something>, etc. Typical implementation:
void CLASS_OF_THIS::makeDeepCopyFromShallowCopy(CopiesMap& copies) { inherited::makeDeepCopyFromShallowCopy(copies); deepCopyField(complex_data_member1, copies); deepCopyField(complex_data_member2, copies); ... }
| copies | A map used by the deep-copy mechanism to keep track of already-copied objects. |
Reimplemented from PLearn::Optimizer.
Definition at line 98 of file OnlineGramNaturalGradientOptimizer.cc.
References cov_eigen_val, cov_eigen_vec, cov_norm_eigen_vec, D, PLearn::deepCopyField(), dot_prod, gradients, gram, PLearn::Optimizer::makeDeepCopyFromShallowCopy(), mu, naturalg, scaled_dot_prod, and U.
{
inherited::makeDeepCopyFromShallowCopy(copies);
deepCopyField(gradients, copies);
deepCopyField(mu, copies);
deepCopyField(gram, copies);
deepCopyField(U, copies);
deepCopyField(D, copies);
deepCopyField(cov_eigen_vec, copies);
deepCopyField(cov_eigen_val, copies);
deepCopyField(cov_norm_eigen_vec, copies);
deepCopyField(dot_prod, copies);
deepCopyField(scaled_dot_prod, copies);
deepCopyField(naturalg, copies);
}

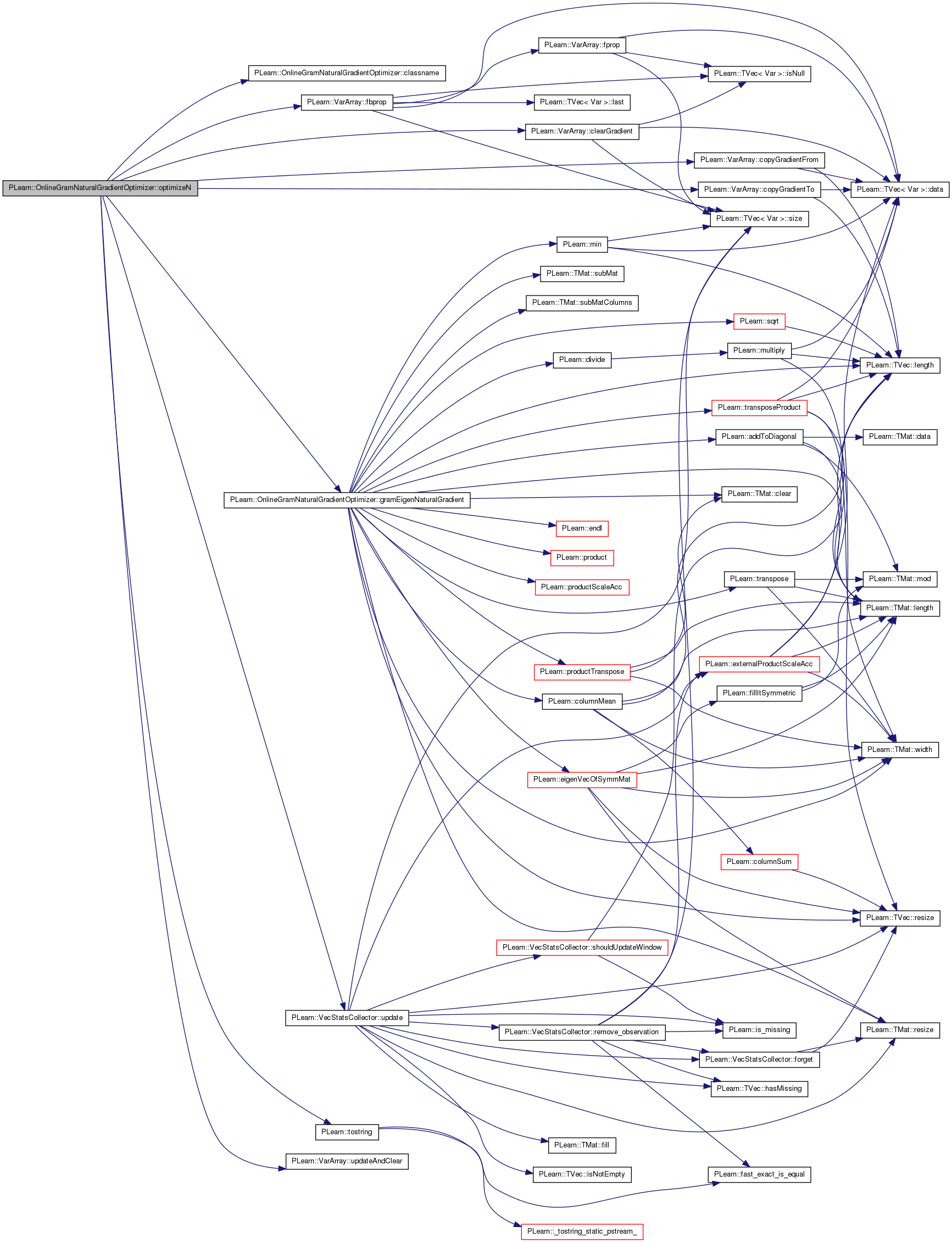
| bool PLearn::OnlineGramNaturalGradientOptimizer::optimizeN | ( | VecStatsCollector & | stats_coll | ) | [virtual] |
Main optimization method, to be defined in subclasses.
Return true iff no further optimization is possible.
Implements PLearn::Optimizer.
Definition at line 144 of file OnlineGramNaturalGradientOptimizer.cc.
References bi, classname(), PLearn::VarArray::clearGradient(), PLearn::VarArray::copyGradientFrom(), PLearn::VarArray::copyGradientTo(), PLearn::Optimizer::cost, PLearn::VarArray::fbprop(), gradients, gramEigenNaturalGradient(), learning_rate, n_optimizeN_calls, naturalg, PLearn::Optimizer::nstages, opt_batch_size, PLearn::Optimizer::params, PLWARNING, PLearn::Optimizer::proppath, PLearn::Optimizer::stage, PLearn::tostring(), PLearn::VecStatsCollector::update(), and PLearn::VarArray::updateAndClear().
{
n_optimizeN_calls++;
if( nstages%opt_batch_size != 0 ) {
PLWARNING("OnlineGramNaturalGradientOptimizer::optimizeN(...) - nstages%opt_batch_size != 0");
}
int stage_max = stage + nstages; // the stage to reach
PP<ProgressBar> pb;
pb = new ProgressBar("Training " + classname() + " from stage "
+ tostring(stage) + " to " + tostring(stage_max), (int)(stage_max-stage)/opt_batch_size );
int initial_stage = stage;
while( stage < stage_max ) {
/*if( bi == 0 )
t0 = clock();*/
// Get the new gradient and append it
params.clearGradient();
proppath.clearGradient();
cost->gradient[0] = -1.0;
proppath.fbprop();
params.copyGradientTo( gradients(bi) );
// End of batch. Compute natural gradient and update parameters.
bi++;
if( bi == opt_batch_size ) {
//t1 = clock();
bi = 0;
gramEigenNaturalGradient();
//t2 = clock();
// set params += -learning_rate * params.gradient
naturalg *= learning_rate;
params.copyGradientFrom( naturalg );
params.updateAndClear();
//t3 = clock();
//cout << double(t1-t0) << " " << double(t2-t1) << " " << double(t3-t2) << endl;
if(pb)
pb->update((stage-initial_stage)/opt_batch_size);
}
stats_coll.update(cost->value);
stage++;
}
return false;
}
Reimplemented from PLearn::Optimizer.
Definition at line 85 of file OnlineGramNaturalGradientOptimizer.h.
Definition at line 121 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by optimizeN().
Definition at line 141 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 138 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 143 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 134 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 146 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 67 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by declareOptions(), and gramEigenNaturalGradient().
Definition at line 126 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), gramEigenNaturalGradient(), makeDeepCopyFromShallowCopy(), and optimizeN().
Definition at line 132 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 66 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by declareOptions(), and optimizeN().
Definition at line 127 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 70 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by declareOptions(), and gramEigenNaturalGradient().
Definition at line 122 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), and gramEigenNaturalGradient().
Definition at line 123 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), and gramEigenNaturalGradient().
Definition at line 118 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), and optimizeN().
Definition at line 150 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), gramEigenNaturalGradient(), makeDeepCopyFromShallowCopy(), and optimizeN().
Definition at line 139 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient().
Definition at line 69 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_(), declareOptions(), and optimizeN().
Definition at line 68 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by declareOptions(), and gramEigenNaturalGradient().
Definition at line 147 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 140 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient().
clock_t PLearn::OnlineGramNaturalGradientOptimizer::t0 [private] |
Definition at line 152 of file OnlineGramNaturalGradientOptimizer.h.
clock_t PLearn::OnlineGramNaturalGradientOptimizer::t1 [private] |
Definition at line 152 of file OnlineGramNaturalGradientOptimizer.h.
clock_t PLearn::OnlineGramNaturalGradientOptimizer::t2 [private] |
Definition at line 152 of file OnlineGramNaturalGradientOptimizer.h.
clock_t PLearn::OnlineGramNaturalGradientOptimizer::t3 [private] |
Definition at line 152 of file OnlineGramNaturalGradientOptimizer.h.
Definition at line 129 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_().
Definition at line 133 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by gramEigenNaturalGradient(), and makeDeepCopyFromShallowCopy().
Definition at line 129 of file OnlineGramNaturalGradientOptimizer.h.
Referenced by build_().
 1.7.4
1.7.4