|
PLearn 0.1
|
|
PLearn 0.1
|
This code is deprecated, use PTester.h and PTester.cc instead. More...
#include <PExperiment.h>
Public Types | |
| typedef Object | inherited |
| typedef map< string, PP < PerformanceEvaluator > > | perf_evaluators_t |
Public Member Functions | |
| PTester () | |
| virtual void | build () |
| Post-constructor. | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual PTester * | deepCopy (CopiesMap &copies) const |
| void | setExperimentDirectory (const PPath &the_expdir) |
| The experiment directory is the directory in which files related to this model are to be saved. | |
| PPath | getExperimentDirectory () const |
| This returns the currently set expdir (see setExperimentDirectory) | |
| virtual void | run () |
| runs the experiment | |
| Vec | perform (bool dont_set_training_set=false) |
| performs the experiment, and returns the global stats specified in statnames If dont_set_training_set is set to true AND the splitter returns only one split, then we *don't* call setTrainingSet() and a forget() on the learner prior to training it: we assume the training set is already set. | |
| PTester () | |
| virtual void | build () |
| Post-constructor. | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual PTester * | deepCopy (CopiesMap &copies) const |
| void | setExperimentDirectory (const PPath &the_expdir) |
| The experiment directory is the directory in which files related to this model are to be saved. | |
| PPath | getExperimentDirectory () const |
| This returns the currently set expdir (see setExperimentDirectory) | |
| TVec< string > | getStatNames () |
| Return the statnames (potentially modified by statmask, if provided). | |
| void | setStatNames (const TVec< string > &the_statnames, bool call_build=true) |
| Set the stat names. | |
| virtual void | run () |
| runs the tester | |
| Vec | perform (bool call_forget=true) |
| Performs the test, and returns the global stats specified in statnames. | |
| Vec | perform1Split (int splitnum, bool call_forget=true) |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Transforms a shallow copy into a deep copy. | |
Static Public Member Functions | |
| static string | _classname_ () |
| Declares name and deepCopy methods. | |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
| static string | _classname_ () |
| Declares name and deepCopy methods. | |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| bool | save_initial_experiment |
| PPath | expdir |
| Path of this experiment's directory in which to save all experiment results (will be created if it does not already exist) | |
| PP< PLearner > | learner |
| PP< Splitter > | splitter |
| TVec< string > | statnames |
| The original statnames option. | |
| bool | report_stats |
| bool | save_stat_collectors |
| bool | save_learners |
| bool | save_initial_learners |
| bool | save_data_sets |
| bool | save_test_outputs |
| bool | save_test_costs |
| bool | provide_learner_expdir |
| VMat | dataset |
| Path of this tester's experiment directory in which to save all tester results (will be created if it does not already exist) | |
| TVec< string > | final_commands |
| PP< VecStatsCollector > | global_template_stats_collector |
| string | save_mode |
| bool | save_initial_tester |
| bool | save_split_stats |
| bool | save_test_names |
| bool | call_forget_in_run |
| TVec< TVec< string > > | statmask |
| PP< VecStatsCollector > | template_stats_collector |
| perf_evaluators_t | perf_evaluators |
| bool | save_test_confidence |
| Whether to save 95% confidence intervals for the test outputs; make sense mostly if 'save_test_outputs' is also true. | |
| bool | should_train |
| Whether or not to train or just test (see 'should_test', below). | |
| bool | should_test |
| Whether to carry out the test at all. | |
| bool | finalize_learner |
| if true, we finalize the learner after training. | |
| bool | enforce_clean_expdir |
| If this option is true, the PTester ensures that the expdir does not already exist when the experiment is started, and gives a PLerror otherwise. | |
| bool | redirect_stdout |
| bool | redirect_stderr |
| bool | parallelize_here |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Protected Member Functions | |
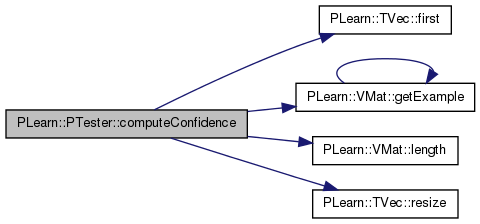
| void | computeConfidence (VMat test_set, VMat confidence) |
| Utility function to compute confidence intervals over a test set and save results in VMat. | |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares this class' options. | |
| static void | declareOptions (OptionList &ol) |
| Declares this class' options. | |
| static void | declareMethods (RemoteMethodMap &rmm) |
| Declare the methods that are remote-callable. | |
Protected Attributes | |
| TVec< string > | statnames_processed |
| The 'real' statnames: these are obtained from 'statnames' by a processing at build time, taking into account the 'statmask' option. | |
| bool | need_to_save_test_names |
| Set to true in perform() when 'save_test_names' is true, in order to remember to save the cost names after setting the learner's training set (since some learners may not have these costs available until they are provided with a training set). | |
| PStream::mode_t | save_mode_ |
| Obtained automatically from the 'save_mode' option. | |
Private Types | |
| typedef Object | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
| void | build_ () |
| This does the actual building. | |
Private Attributes | |
| bool | reloaded |
This code is deprecated, use PTester.h and PTester.cc instead.
Definition at line 53 of file PExperiment.h.
| typedef Object PLearn::PTester::inherited |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 57 of file PExperiment.h.
typedef Object PLearn::PTester::inherited [private] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| typedef map<string, PP<PerformanceEvaluator> > PLearn::PTester::perf_evaluators_t |
| PLearn::PTester::PTester | ( | ) |
Definition at line 72 of file PExperiment.cc.
: report_stats(true), save_initial_experiment(true), save_stat_collectors(true), save_learners(true), save_initial_learners(false), save_data_sets(false), save_test_outputs(false), save_test_costs(false), provide_learner_expdir(false) {}
| PLearn::PTester::PTester | ( | ) |
| string PLearn::PTester::_classname_ | ( | ) | [static] |
Declares name and deepCopy methods.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| static string PLearn::PTester::_classname_ | ( | ) | [static] |
Declares name and deepCopy methods.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| OptionList & PLearn::PTester::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| static OptionList& PLearn::PTester::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| RemoteMethodMap & PLearn::PTester::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| static RemoteMethodMap& PLearn::PTester::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| static Object* PLearn::PTester::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| Object * PLearn::PTester::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| static void PLearn::PTester::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| StaticInitializer PTester::_static_initializer_ & PLearn::PTester::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| void PLearn::PTester::build | ( | ) | [virtual] |
Post-constructor.
The normal implementation should call simply inherited::build(), then this class's build_(). This method should be callable again at later times, after modifying some option fields to change the "architecture" of the object.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 151 of file PExperiment.cc.
References PLearn::Object::build(), and build_().
{
inherited::build();
build_();
}
| virtual void PLearn::PTester::build | ( | ) | [virtual] |
Post-constructor.
The normal implementation should call simply inherited::build(), then this class's build_(). This method should be callable again at later times, after modifying some option fields to change the "architecture" of the object.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| void PLearn::PTester::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| void PLearn::PTester::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 138 of file PExperiment.cc.
References expdir, PLearn::force_mkdir(), PLearn::pathexists(), and PLERROR.
Referenced by build().
{
if(expdir!="")
{
if(pathexists(expdir))
PLERROR("Directory (or file) %s already exists. First move it out of the way.",expdir.c_str());
if(!force_mkdir(expdir))
PLERROR("In PTester Could not create experiment directory %s",expdir.c_str());
expdir = abspath(expdir);
}
}

| string PLearn::PTester::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| virtual string PLearn::PTester::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Utility function to compute confidence intervals over a test set and save results in VMat.
Definition at line 921 of file PTester.cc.
References PLearn::TVec< T >::first(), PLearn::VMat::getExample(), i, j, PLearn::VMat::length(), n, PLASSERT, and PLearn::TVec< T >::resize().
{
PLASSERT(learner);
if (!confidence)
return;
PP<ProgressBar> pb;
const int n = test_set.length();
if (learner->report_progress)
pb = new ProgressBar("Computing Confidence Intervals", n);
Vec input, target, output(learner->outputsize());
TVec< pair<real,real> > intervals;
Vec intervals_real;
real weight;
for (int i=0 ; i<n ; ++i) {
if (pb)
pb->update(i);
test_set.getExample(i, input, target, weight);
learner->computeOutput(input,output);
learner->computeConfidenceFromOutput(input,output,0.95,intervals);
intervals_real.resize(2*intervals.size());
for (int j=0 ; j<intervals.size() ; ++j) {
intervals_real[2*j] = intervals[j].first;
intervals_real[2*j+1] = intervals[j].second;
}
confidence->putOrAppendRow(i,intervals_real);
}
}
| void PLearn::PTester::declareMethods | ( | RemoteMethodMap & | rmm | ) | [static, protected] |
Declare the methods that are remote-callable.
Reimplemented from PLearn::Object.
Definition at line 313 of file PTester.cc.
References PLearn::declareMethod(), and PLearn::RemoteMethodMap::inherited().
{
// Insert a backpointer to remote methods; note that this
// different than for declareOptions()
rmm.inherited(inherited::_getRemoteMethodMap_());
declareMethod(
rmm, "perform", &PTester::perform,
(BodyDoc("Performs the test, and returns the global stats specified in statnames.\n"
"If 'call_forget' is set to false then the call to setTrainingSet()\n"
"won't call forget and build. This is useful for continuation of an\n"
"incremental training (such as after increasing the number of epochs\n"
"(nstages) ), or generally when trying different option values that\n"
"don't require the learning to be restarted from scratch. However\n"
"call_forget will be forced to true (even if passed as false) if the\n"
"splitter returns more than one split.\n"),
ArgDoc ("call_forget", "Whether forget() should be called in setTrainingSet()."),
RetDoc ("Vector of test statistics corresponding to the requested statnames")));
declareMethod(
rmm, "perform1Split", &PTester::perform1Split,
(BodyDoc("Performs train/test for one split, returns splitres."),
ArgDoc ("splitnum","Split number on which to perform train/test"),
ArgDoc ("call_forget","Whether forget() should be called in setTrainingSet()."),
RetDoc ("Vector of test statistics corresponding to the requested statnames")));
declareMethod(
rmm, "getStatNames", &PTester::getStatNames,
(BodyDoc("Return the statnames (potentially modified by statmask, if provided);\n"
"see the 'statnames' and 'statmask' options."),
RetDoc ("Name of computed statistics.")));
declareMethod(
rmm, "setExperimentDirectory", &PTester::setExperimentDirectory,
(BodyDoc("The experiment directory is the directory in which files related to\n"
"this model are to be saved. If it is an empty string, it is understood\n"
"to mean that the user doesn't want any file created by this learner.\n"),
ArgDoc ("expdir", "Directory name where experimental results should be saved")));
declareMethod(
rmm, "getExperimentDirectory", &PTester::getExperimentDirectory,
(BodyDoc("Return the currently-set experiment directory (see setExperimentDirectory)."),
RetDoc ("Current expdir.")));
}

| void PLearn::PTester::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares this class' options.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 92 of file PExperiment.cc.
References PLearn::OptionBase::buildoption, PLearn::declareOption(), PLearn::Object::declareOptions(), expdir, learner, provide_learner_expdir, report_stats, save_data_sets, save_initial_experiment, save_initial_learners, save_learners, save_stat_collectors, save_test_costs, save_test_outputs, splitter, and statnames.
{
declareOption(ol, "expdir", &PTester::expdir, OptionBase::buildoption,
"Path of this experiment's directory in which to save all experiment results.\n"
"The directory will be created if it does not already exist.\n"
"If this is an empty string, no directory is created and no output file is generated.\n");
declareOption(ol, "learner", &PTester::learner, OptionBase::buildoption,
"The learner to train/test.\n"
"learner.train_set will be used as the dataset for this experiment\n"
"(you may omit learner.train_set if your splitter is an ExplicitSplitter)");
declareOption(ol, "splitter", &PTester::splitter, OptionBase::buildoption,
"The splitter to use to generate one or several train/test tuples from the dataset.");
declareOption(ol, "statnames", &PTester::statnames, OptionBase::buildoption,
"A list of global statistics we are interested in.\n"
"These are strings of the form S1[S2[dataset.cost_name]] where:\n"
" - dataset is train or test1 or test2 ... (train being \n"
" the first dataset in a split, test1 the second, ...) \n"
" - cost_name is one of the training or test cost names (depending on dataset) understood \n"
" by the underlying learner (see its getTrainCostNames and getTestCostNames methods) \n"
" - S1 and S2 are a statistic, i.e. one of: E (expectation), V(variance), MIN, MAX, STDDEV, ... \n"
" S2 is computed over the samples of a given dataset split. S1 is over the splits. \n");
declareOption(ol, "report_stats", &PTester::report_stats, OptionBase::buildoption,
"If true, the computed global statistics specified in statnames will be saved in global_stats.pmat \n"
"and the corresponding per-split statistics will be saved in split_stats.pmat \n"
"For reference, all cost names (as given by the learner's getTrainCostNames() and getTestCostNames() ) \n"
"will be reported in files train_cost_names.txt and test_cost_names.txt");
declareOption(ol, "save_initial_experiment", &PTester::save_initial_experiment, OptionBase::buildoption,
"If true, this PTester object will be saved in its initial state in experiment.psave \n"
"Thus if the initial .plearn file gets lost, or modified, we can always see what this experiment was.\n");
declareOption(ol, "save_stat_collectors", &PTester::save_stat_collectors, OptionBase::buildoption,
"If true, stat collectors for split#k will be saved in Split#k/train_stats.psave and Split#k/test#i_stats.psave");
declareOption(ol, "save_learners", &PTester::save_learners, OptionBase::buildoption,
"If true, the final trained learner for split#k will be saved in Split#k/final_learner.psave");
declareOption(ol, "save_initial_learners", &PTester::save_initial_learners, OptionBase::buildoption,
"If true, the initial untrained learner for split#k (just after forget() has been called) will be saved in Split#k/initial_learner.psave");
declareOption(ol, "save_data_sets", &PTester::save_data_sets, OptionBase::buildoption,
"If true, the data set generated for split #k will be saved as Split#k/training_set.psave Split#k/test1_set.psave ...");
declareOption(ol, "save_test_outputs", &PTester::save_test_outputs, OptionBase::buildoption,
"If true, the outputs of the test for split #k will be saved in Split#k/test#i_outputs.pmat");
declareOption(ol, "save_test_costs", &PTester::save_test_costs, OptionBase::buildoption,
"If true, the costs of the test for split #k will be saved in Split#k/test#i_costs.pmat");
declareOption(ol, "provide_learner_expdir", &PTester::provide_learner_expdir, OptionBase::buildoption,
"If true, each learner to be trained will have its experiment directory set to Split#k/LearnerExpdir/");
inherited::declareOptions(ol);
}
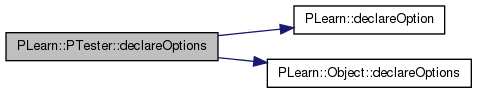
| static void PLearn::PTester::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares this class' options.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| static const PPath& PLearn::PTester::declaringFile | ( | ) | [inline, static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 180 of file PTester.h.
{ return expdir; }
| static const PPath& PLearn::PTester::declaringFile | ( | ) | [inline, static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 107 of file PExperiment.h.
{ return expdir; }
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| PPath PLearn::PTester::getExperimentDirectory | ( | ) | const [inline] |
This returns the currently set expdir (see setExperimentDirectory)
Definition at line 116 of file PExperiment.h.
{ return expdir; }
| PPath PLearn::PTester::getExperimentDirectory | ( | ) | const [inline] |
| virtual OptionList& PLearn::PTester::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| OptionList & PLearn::PTester::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| OptionMap & PLearn::PTester::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| virtual OptionMap& PLearn::PTester::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| RemoteMethodMap & PLearn::PTester::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 90 of file PExperiment.cc.
| virtual RemoteMethodMap& PLearn::PTester::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
| TVec< string > PLearn::PTester::getStatNames | ( | ) |
Return the statnames (potentially modified by statmask, if provided).
Definition at line 964 of file PTester.cc.
{
return statnames_processed;
}
| void PLearn::PTester::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Transforms a shallow copy into a deep copy.
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 1025 of file PTester.cc.
References PLearn::deepCopyField().
{
inherited::makeDeepCopyFromShallowCopy(copies);
deepCopyField(statnames, copies);
deepCopyField(statnames_processed, copies);
deepCopyField(dataset, copies);
deepCopyField(final_commands, copies);
deepCopyField(global_template_stats_collector, copies);
deepCopyField(learner, copies);
deepCopyField(splitter, copies);
deepCopyField(statmask, copies);
deepCopyField(template_stats_collector, copies);
deepCopyField(perf_evaluators, copies);
}

performs the experiment, and returns the global stats specified in statnames If dont_set_training_set is set to true AND the splitter returns only one split, then we *don't* call setTrainingSet() and a forget() on the learner prior to training it: we assume the training set is already set.
This is useful for continuation of an incremental training (such as after increasing the number of epochs (nstages) ).
Definition at line 261 of file PExperiment.cc.
References PLearn::append_slash(), PLearn::OptionBase::buildoption, PLearn::StatSpec::costindex, dataset, expdir, PLearn::StatSpec::intstat, PLearn::join(), learner, PLearn::TVec< T >::length(), PLERROR, provide_learner_expdir, report_stats, PLearn::Object::save(), save_data_sets, save_initial_experiment, save_initial_learners, save_learners, save_stat_collectors, save_test_costs, save_test_outputs, PLearn::saveStringInFile(), PLearn::StatSpec::setnum, PLearn::TVec< T >::size(), slash, splitter, statnames, PLearn::TVec< T >::subVec(), and PLearn::tostring().
Referenced by run().
{
if(!learner)
PLERROR("No learner specified for PTester.");
if(!splitter)
PLERROR("No splitter specified for PTester");
// get initial data set.
VMat dataset = learner->getTrainingSet();
if(expdir!="")
{
// Save this experiment description in the expdir (buildoptions only)
if(save_initial_experiment)
PLearn::save(append_slash(expdir)+"experiment.psave", *this, OptionBase::buildoption);
}
splitter->setDataSet(dataset);
int nsplits = splitter->nsplits();
TVec<string> testcostnames = learner->getTestCostNames();
TVec<string> traincostnames = learner->getTrainCostNames();
int nsets = splitter->nSetsPerSplit();
int nstats = statnames.length();
// Stats collectors for individual sets of a split:
TVec< PP<VecStatsCollector> > stcol(nsets);
for(int setnum=0; setnum<nsets; setnum++)
stcol[setnum] = new VecStatsCollector();
PP<VecStatsCollector> train_stats = stcol[0];
learner->setTrainStatsCollector(train_stats);
// Global stats collector
PP<VecStatsCollector> global_statscol = new VecStatsCollector();
// Stat specs
TVec<StatSpec> statspecs(nstats);
for(int k=0; k<nstats; k++)
statspecs[k].init(statnames[k],learner);
// int traincostsize = traincostnames.size();
int testcostsize = testcostnames.size();
int outputsize = learner->outputsize();
VMat global_stats_vm; // the vmat in which to save global result stats specified in statnames
VMat split_stats_vm; // the vmat in which to save per split result stats
if(expdir!="" && report_stats)
{
saveStringInFile(expdir+slash+"train_cost_names.txt", join(traincostnames,"\n")+"\n");
saveStringInFile(expdir+slash+"test_cost_names.txt", join(testcostnames,"\n")+"\n");
global_stats_vm = new FileVMatrix(expdir+slash+"global_stats.pmat", 1, nstats);
for(int k=0; k<nstats; k++)
global_stats_vm->declareField(k,statspecs[k].statName());
global_stats_vm->saveFieldInfos();
split_stats_vm = new FileVMatrix(expdir+slash+"split_stats.pmat", nsplits, 1+nstats);
split_stats_vm->declareField(0,"splitnum");
for(int k=0; k<nstats; k++)
split_stats_vm->declareField(k+1,statspecs[k].intStatName());
split_stats_vm->saveFieldInfos();
}
for(int splitnum=0; splitnum<nsplits; splitnum++)
{
string splitdir;
if(expdir!="")
splitdir = append_slash(expdir)+"Split"+tostring(splitnum)+slash;
TVec<VMat> dsets = splitter->getSplit(splitnum);
VMat trainset = dsets[0];
if(splitdir!="" && save_data_sets)
PLearn::save(splitdir+"training_set.psave",trainset);
if(splitdir!="" && provide_learner_expdir)
learner->setExperimentDirectory(splitdir+"LearnerExpdir"+slash);
if(!dont_set_training_set || nsplits>1)
learner->setTrainingSet(trainset); // also calls forget...
if(splitdir!="" && save_initial_learners)
PLearn::save(splitdir+"initial_learner.psave",learner);
train_stats->forget();
learner->train();
train_stats->finalize();
if(save_stat_collectors)
PLearn::save(splitdir+"train_stats.psave",train_stats);
if(save_learners)
PLearn::save(splitdir+"final_learner.psave",learner);
for(int setnum=1; setnum<dsets.length(); setnum++)
{
VMat testset = dsets[setnum];
PP<VecStatsCollector> test_stats = stcol[setnum];
string setname = "test"+tostring(setnum);
if(splitdir!="" && save_data_sets)
PLearn::save(splitdir+setname+"_set.psave",testset);
VMat test_outputs;
VMat test_costs;
if(save_test_outputs)
test_outputs = new FileVMatrix(splitdir+setname+"_outputs.pmat",0,outputsize);
if(save_test_costs)
test_costs = new FileVMatrix(splitdir+setname+"_costs.pmat",0,testcostsize);
test_stats->forget();
learner->test(testset, test_stats, test_outputs, test_costs);
test_stats->finalize();
if(save_stat_collectors)
PLearn::save(splitdir+setname+"_stats.psave",test_stats);
}
Vec splitres(1+nstats);
splitres[0] = splitnum;
for(int k=0; k<nstats; k++)
{
StatSpec& sp = statspecs[k];
splitres[k+1] = stcol[sp.setnum]->getStats(sp.costindex).getStat(sp.intstat);
}
if(split_stats_vm)
split_stats_vm->appendRow(splitres);
global_statscol->update(splitres.subVec(1,nstats));
}
Vec global_result(nstats);
global_statscol->finalize();
for(int k=0; k<nstats; k++)
global_result[k] = global_statscol->getStats(k).getStat(statspecs[k].extstat);
if(global_stats_vm)
global_stats_vm->appendRow(global_result);
return global_result;
}
Performs the test, and returns the global stats specified in statnames.
If 'call_forget' is set to false then the call to setTrainingSet() won't call forget and build. This is useful for continuation of an incremental training (such as after increasing the number of epochs (nstages) ), or generally when trying different option values that don't require the learning to be restarted from scratch. However call_forget will be forced to true (even if passed as false) if the splitter returns more than one split.
Returns a vector of test statistics corresponding to the requested statnames
Definition at line 504 of file PTester.cc.
References PLearn::TVec< T >::deepCopy(), PLearn::force_mkdir(), PLearn::StatSpec::intstatname, PLearn::TVec< T >::isEmpty(), PLearn::join(), PLearn::left(), PLearn::TVec< T >::length(), PLearn::VMat::length(), MISSING_VALUE, PLERROR, PLWARNING, PLearn::TMat< T >::resize(), PLearn::right(), PLearn::save(), PLearn::saveStringInFile(), PLearn::StatSpec::setnum, PLearn::TVec< T >::size(), PLearn::split_on_first(), and PLearn::tostring().
{
if (!learner)
PLERROR("PTester::perform1Split : No learner specified for PTester.");
if (!splitter)
PLERROR("PTester::perform1Split : No splitter specified for PTester");
const int nstats = statnames_processed.length();
const int nsets = splitter->nSetsPerSplit();
// Stats collectors for individual sets of a split:
TVec< PP<VecStatsCollector> > stcol(nsets);
for (int setnum = 0; setnum < nsets; setnum++)
{
if (template_stats_collector)
{
CopiesMap copies;
stcol[setnum] = template_stats_collector->deepCopy(copies);
}
else
stcol[setnum] = new VecStatsCollector();
}
// Stat specs
TVec<StatSpec> statspecs(nstats);
for(int k = 0; k < nstats; k++)
{
statspecs[k].init(statnames_processed[k]);
}
PPath splitdir;
bool is_splitdir = false;
if (!expdir.isEmpty())
{
splitdir = expdir / ("Split" + tostring(splitnum));
is_splitdir = true;
}
TVec<VMat> dsets = splitter->getSplit(splitnum);
TVec<string> testcostnames;
if (should_train) {
VMat trainset = dsets[0];
if (is_splitdir && save_data_sets)
PLearn::save(splitdir / "training_set.vmat", trainset);
if (provide_learner_expdir)
{
if (is_splitdir)
learner->setExperimentDirectory(splitdir / "LearnerExpdir/");
else
learner->setExperimentDirectory("");
}
learner->setTrainingSet(trainset, call_forget);
testcostnames = learner->getTestCostNames();
TVec<string> traincostnames = learner->getTrainCostNames();
PP<VecStatsCollector> train_stats = stcol[0];
train_stats->setFieldNames(traincostnames);
train_stats->build();
train_stats->forget();
learner->setTrainStatsCollector(train_stats);
if (need_to_save_test_names) {
// Now that the learner has a training set, we can be sure the
// cost names can be saved.
saveStringInFile(expdir / "train_cost_names.txt", join(traincostnames, "\n") + "\n");
saveStringInFile(expdir / "test_cost_names.txt", join(testcostnames, "\n") + "\n");
need_to_save_test_names = false;
}
if (dsets.size() > 1)
learner->setValidationSet(dsets[1]);
if (is_splitdir && save_initial_learners)
PLearn::save(splitdir / "initial_learner.psave", learner);
train_stats->forget();
learner->train();
if(finalize_learner)
learner->finalize();
train_stats->finalize();
if (is_splitdir)
{
if (save_stat_collectors)
PLearn::save(splitdir / "train_stats.psave", train_stats);
if (save_learners)
PLearn::save(splitdir / "final_learner.psave", learner, save_mode_);
}
}
else
learner->build();
// This needs to be after the SetTrainingSet() / build() call to the
// learner.
const int outputsize = learner->outputsize();
// perf_eval_costs[setnum][perf_evaluator_name][costname] will contain value
// of the given cost returned by the given perf_evaluator on the given setnum
TVec< map<string, map<string, real> > > perf_eval_costs(dsets.length());
if (testcostnames.isEmpty())
testcostnames = learner->getTestCostNames();
for (int setnum = 1; setnum < nsets; setnum++) {
stcol[setnum]->setFieldNames(testcostnames);
stcol[setnum]->build();
stcol[setnum]->forget();
}
// Perform the test if required
if (should_test)
{
for (int setnum = 1; setnum < dsets.length(); setnum++)
{
VMat testset = dsets[setnum];
VMat test_outputs;
VMat test_costs;
VMat test_confidence;
PP<VecStatsCollector> test_stats = stcol[setnum];
const string setname = "test" + tostring(setnum);
if (is_splitdir && save_data_sets)
PLearn::save(splitdir / (setname + "_set.vmat"), testset);
// QUESTION Why is this done so late? Can't it be moved
// somewhere earlier? At least before the save_data_sets?
if (is_splitdir)
force_mkdir(splitdir);
if (is_splitdir && save_test_outputs)
test_outputs = new FileVMatrix(splitdir / (setname + "_outputs.pmat"),
0, learner->getOutputNames());
else if (!perf_evaluators.empty())
{
// We don't want to save test outputs to disk, but we
// need them for pef_evaluators. So let's store them in
// a MemoryVMatrix
Mat data(testset.length(), outputsize);
data.resize(0, outputsize);
test_outputs = new MemoryVMatrix(data);
test_outputs->declareFieldNames(learner->getOutputNames());
}
if (is_splitdir)
{
if (save_test_costs)
test_costs = new FileVMatrix(splitdir / (setname + "_costs.pmat"),
0, learner->getTestCostNames());
if (save_test_confidence)
test_confidence = new FileVMatrix(splitdir / (setname + "_confidence.pmat"),
0, 2 * outputsize);
}
test_stats->forget();
if (testset->length() == 0)
PLWARNING("PTester:: test set %s is of length 0, costs will be set to -1",
setname.c_str());
// Before each test set, reset the internal state of the learner
learner->resetInternalState();
learner->test(testset, test_stats, test_outputs, test_costs);
//if (reset_stats)
test_stats->finalize();
if (is_splitdir && save_stat_collectors)
PLearn::save(splitdir / (setname + "_stats.psave"), test_stats);
perf_evaluators_t::iterator it = perf_evaluators.begin();
const perf_evaluators_t::iterator itend = perf_evaluators.end();
while (it != itend)
{
PPath perf_eval_dir;
if (is_splitdir)
perf_eval_dir = splitdir / setname / ("perfeval_" + it->first);
Vec perf_costvals = it->second->evaluatePerformance(learner, testset, test_outputs, perf_eval_dir);
TVec<string> perf_costnames = it->second->getCostNames();
if (perf_costvals.length()!=perf_costnames.length())
PLERROR("vector of costs returned by performance evaluator differ in size with its vector of costnames");
map<string, real>& costmap = perf_eval_costs[setnum][it->first];
for (int costi = 0; costi < perf_costnames.length(); costi++)
costmap[perf_costnames[costi]] = perf_costvals[costi];
++it;
}
computeConfidence(testset, test_confidence);
}
}
Vec splitres(1 + nstats);
splitres[0] = splitnum;
for (int k = 0; k < nstats; k++)
{
// If we ask for a test-set that's beyond what's currently
// available, OR we are asking for test-statistics in
// train-only mode, then the statistic is MISSING_VALUE.
StatSpec& sp = statspecs[k];
if (sp.setnum>=stcol.length() ||
(! should_test && sp.setnum > 0))
{
splitres[k+1] = MISSING_VALUE;
}
else
{
string left, right;
split_on_first(sp.intstatname, ".",left,right);
if (right != "" && perf_evaluators.find(left) != perf_evaluators.end())
{
// looks like a cost from a performance evaluator
map<string, real>& costmap = perf_eval_costs[sp.setnum][left];
if (costmap.find(right) == costmap.end())
PLERROR("No cost named %s appears to be returned by evaluator %s",
right.c_str(), left.c_str());
splitres[k+1] = costmap[right];
}
else
// must be a cost from a stats collector
splitres[k+1] = stcol[sp.setnum]->getStat(sp.intstatname);
}
}
return splitres;
}
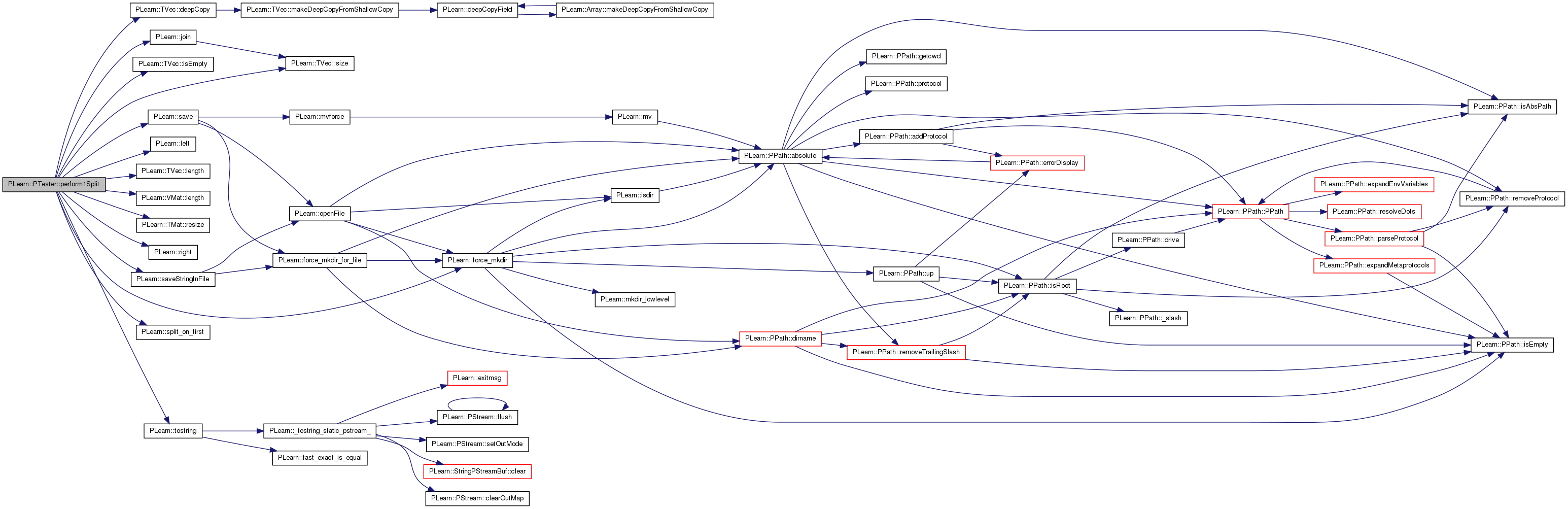
| virtual void PLearn::PTester::run | ( | ) | [virtual] |
runs the tester
Reimplemented from PLearn::Object.
| void PLearn::PTester::run | ( | ) | [virtual] |
runs the experiment
Reimplemented from PLearn::Object.
Definition at line 157 of file PExperiment.cc.
References perform().
{
perform(false);
}
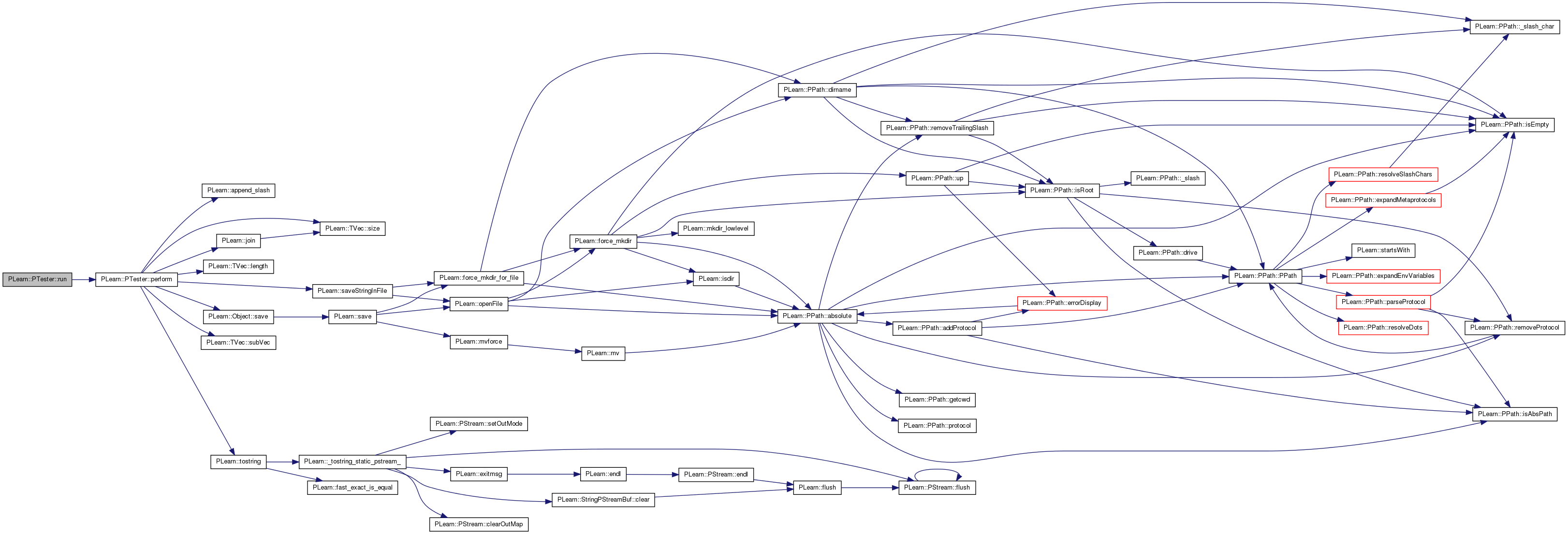
| void PLearn::PTester::setExperimentDirectory | ( | const PPath & | the_expdir | ) |
The experiment directory is the directory in which files related to this model are to be saved.
If it is an empty string, it is understood to mean that the user doesn't want any file created by this learner.
Definition at line 249 of file PExperiment.cc.
References expdir, PLearn::force_mkdir(), and PLERROR.
{
if(the_expdir=="")
expdir = "";
else
{
if(!force_mkdir(the_expdir))
PLERROR("In PTester::setExperimentDirectory Could not create experiment directory %s",the_expdir.c_str());
expdir = abspath(the_expdir);
}
}
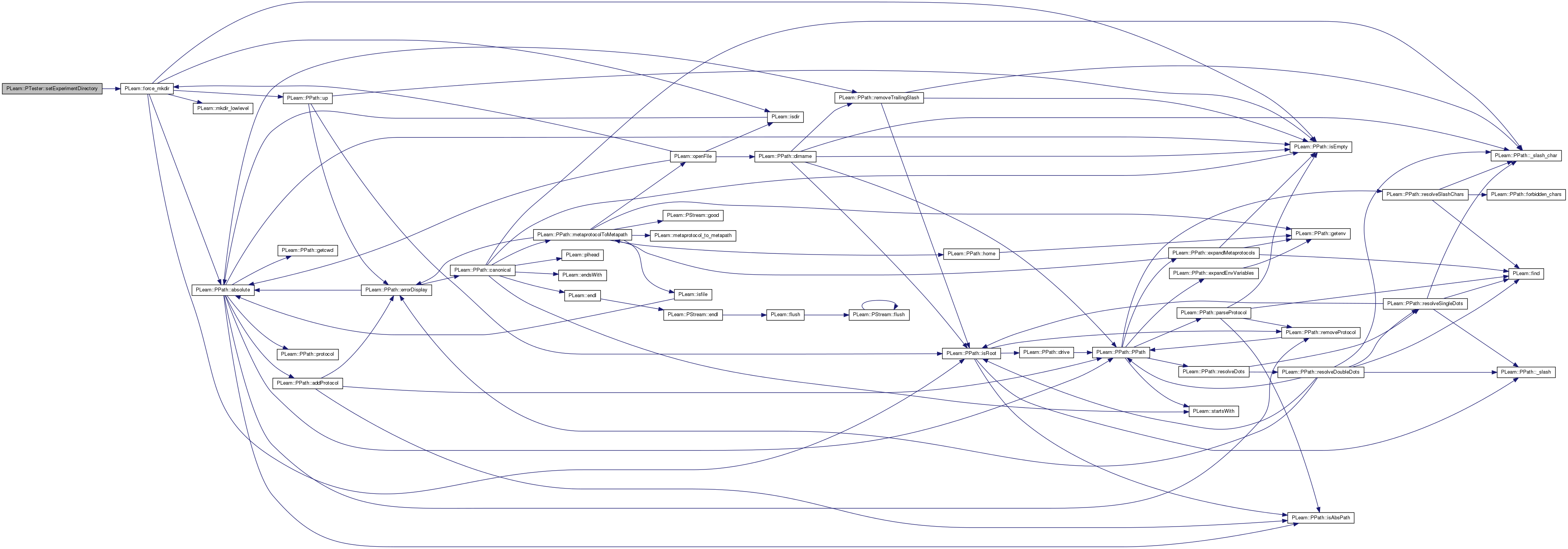
| void PLearn::PTester::setExperimentDirectory | ( | const PPath & | the_expdir | ) |
The experiment directory is the directory in which files related to this model are to be saved.
If it is an empty string, it is understood to mean that the user doesn't want any file created by this learner.
Set the stat names.
The vector 'statnames' is copied. By default, the object is re-built, but this can be disabled by setting 'call_build' to false.
Definition at line 952 of file PTester.cc.
References PLearn::TVec< T >::length().

static StaticInitializer PLearn::PTester::_static_initializer_ [static] |
Reimplemented from PLearn::Object.
Reimplemented in PLearn::SecondIterationTester.
Definition at line 107 of file PExperiment.h.
If this option is true, the PTester ensures that the expdir does not already exist when the experiment is started, and gives a PLerror otherwise.
This is the usual and traditional default behavior for PTester. However, in some contexts, one KNOWS that the expdir is brand new (e.g. generated by plargs.expdir in a PTester), and might contain some precomputed results that are being generated as the model is loaded, so it is not empty. In those contexts, it makes sense to allow this option to be false.
Path of this experiment's directory in which to save all experiment results (will be created if it does not already exist)
Definition at line 66 of file PExperiment.h.
Referenced by build_(), declareOptions(), perform(), and setExperimentDirectory().
| TVec<string> PLearn::PTester::final_commands |
Definition at line 67 of file PExperiment.h.
Referenced by declareOptions(), and perform().
bool PLearn::PTester::need_to_save_test_names [protected] |
Definition at line 78 of file PExperiment.h.
Referenced by declareOptions(), and perform().
bool PLearn::PTester::reloaded [private] |
Definition at line 70 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 75 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 56 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 74 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 73 of file PExperiment.h.
Referenced by declareOptions(), and perform().
| string PLearn::PTester::save_mode |
PStream::mode_t PLearn::PTester::save_mode_ [protected] |
Definition at line 72 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 77 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 76 of file PExperiment.h.
Referenced by declareOptions(), and perform().
Definition at line 68 of file PExperiment.h.
Referenced by declareOptions(), and perform().
| TVec<TVec<string> > PLearn::PTester::statmask |
| TVec< string > PLearn::PTester::statnames |
The original statnames option.
It is private because it is safer to access stats from getStatNames, since the 'statmask' option may modify the stats.
Definition at line 69 of file PExperiment.h.
Referenced by declareOptions(), and perform().
TVec<string> PLearn::PTester::statnames_processed [protected] |
 1.7.4
1.7.4