|
PLearn 0.1
|
|
PLearn 0.1
|
Subclass of Molecule, plus standard devs of points' positions and features. More...
#include <MoleculeTemplate.h>
Public Member Functions | |
| MoleculeTemplate (int the_class_label=-1) | |
| Default constructor. | |
| MoleculeTemplate (const PPath &filename, int the_class_label=-1) | |
| Constructor from files. | |
| MoleculeTemplate (const Molecule &molecule, const Vec &the_geom_dev=Vec(), const Mat &the_feat_dev=Mat(), int the_class_label=-1) | |
| Constructor from parent. | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual MoleculeTemplate * | deepCopy (CopiesMap &copies) const |
| virtual void | build () |
| Post-constructor. | |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Transforms a shallow copy into a deep copy. | |
Static Public Member Functions | |
| static string | _classname_ () |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| Vec | geom_dev |
| ### declare public option fields (such as build options) here | |
| Mat | feat_dev |
| standard deviations of each chemical property | |
| int | class_label |
| class label, 0 for inactive, 1 for active, -1 for uninitialized | |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Protected Member Functions | |
| virtual void | readFromAMATFile (const PPath &filename) |
| Override inherited function to read also deviation info, if able. | |
| virtual void | writeToAMATFile (const PPath &filename) |
| Override inherited function to write also deviation info. | |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares the class options. | |
Private Types | |
| typedef Molecule | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
Subclass of Molecule, plus standard devs of points' positions and features.
There is only one geometric standard deviation per point (since space dimenstions are equivalent), there is one chemical standard deviation per chemical feature on every point (stored in the same order as the corresponding feature value).
Definition at line 59 of file MoleculeTemplate.h.
typedef Molecule PLearn::MoleculeTemplate::inherited [private] |
Reimplemented from PLearn::Molecule.
Definition at line 61 of file MoleculeTemplate.h.
| PLearn::MoleculeTemplate::MoleculeTemplate | ( | int | the_class_label = -1 | ) |
Default constructor.
Definition at line 59 of file MoleculeTemplate.cc.
: class_label( the_class_label ) { }
Constructor from files.
Definition at line 64 of file MoleculeTemplate.cc.
References build(), and PLearn::Molecule::readFromFile().
: class_label( the_class_label ) { readFromFile( filename ); build(); }
| PLearn::MoleculeTemplate::MoleculeTemplate | ( | const Molecule & | molecule, |
| const Vec & | the_geom_dev = Vec(), |
||
| const Mat & | the_feat_dev = Mat(), |
||
| int | the_class_label = -1 |
||
| ) |
Constructor from parent.
Definition at line 72 of file MoleculeTemplate.cc.
: inherited( molecule ), geom_dev( the_geom_dev ), feat_dev( the_feat_dev ), class_label( the_class_label ) { // build(); }
| string PLearn::MoleculeTemplate::_classname_ | ( | ) | [static] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| OptionList & PLearn::MoleculeTemplate::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| RemoteMethodMap & PLearn::MoleculeTemplate::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| Object * PLearn::MoleculeTemplate::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| StaticInitializer MoleculeTemplate::_static_initializer_ & PLearn::MoleculeTemplate::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| void PLearn::MoleculeTemplate::build | ( | ) | [virtual] |
Post-constructor.
The normal implementation should call simply inherited::build(), then this class's build_(). This method should be callable again at later times, after modifying some option fields to change the "architecture" of the object.
Reimplemented from PLearn::Molecule.
Definition at line 84 of file MoleculeTemplate.cc.
References build_().
Referenced by MoleculeTemplate().
{
inherited::build();
build_();
}

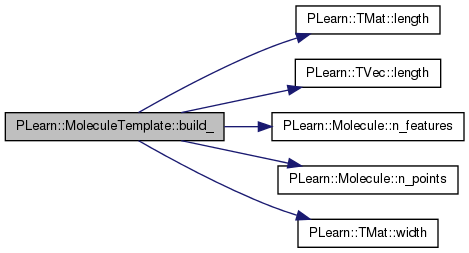
| void PLearn::MoleculeTemplate::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::Molecule.
Definition at line 186 of file MoleculeTemplate.cc.
References feat_dev, geom_dev, PLearn::TMat< T >::length(), PLearn::TVec< T >::length(), PLearn::Molecule::n_features(), PLearn::Molecule::n_points(), PLERROR, PLWARNING, and PLearn::TMat< T >::width().
Referenced by build().
{
// Size check
int geom_dev_length = geom_dev.length();
int feat_dev_length = feat_dev.length();
int feat_dev_width = feat_dev.width();
// TODO: resize if empty?
if( geom_dev_length == 0 )
{
PLWARNING( "MoleculeTemplate::build_ - geom_dev.length() == 0,\n"
"resizing to n_points() (%d), and filling with"
" 1's.\n", n_points() );
geom_dev = Vec( n_points(), 1 );
}
else if( geom_dev_length != n_points() )
PLERROR( "MoleculeTemplate::build_ - geom_dev.length() should be equal"
" to\n"
"n_points() (%d != %d).\n",
geom_dev_length, n_points() );
if( feat_dev_length == 0 && feat_dev_width == 0 )
{
PLWARNING( "MoleculeTemplate::build_ - feat_dev.length() == 0 and\n"
"feat_dev.width() == 0. Resizing to n_points() ×"
" n_features()\n"
"(%d × %d), and filling with 1's.\n",
n_points(), n_features() );
feat_dev = Mat( n_points(), n_features(), 1 );
}
else if( feat_dev_length != n_points() )
PLERROR( "MoleculeTemplate::build_ - feat_dev.length() should be equal"
" to\n"
"n_points() (%d != %d).\n",
feat_dev_length, n_points() );
else if( feat_dev_width != n_features() )
PLERROR( "MoleculeTemplate::build_ - feat_dev.width() should be equal"
" to\n"
"n_features() (%d != %d).\n", feat_dev_width, n_features() );
}

| string PLearn::MoleculeTemplate::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| void PLearn::MoleculeTemplate::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares the class options.
Reimplemented from PLearn::Molecule.
Definition at line 164 of file MoleculeTemplate.cc.
References PLearn::OptionBase::buildoption, class_label, PLearn::declareOption(), feat_dev, and geom_dev.
{
// declareOption(ol, "myoption", &MoleculeTemplate::myoption, OptionBase::buildoption,
// "Help text describing this option");
declareOption(ol, "geom_dev", &MoleculeTemplate::geom_dev,
OptionBase::buildoption,
"Standard deviations of the geometrical distance");
declareOption(ol, "feat_dev", &MoleculeTemplate::feat_dev,
OptionBase::buildoption,
"Standard deviations of each chemical property");
declareOption(ol, "class_label", &MoleculeTemplate::class_label,
OptionBase::buildoption,
"Class label (0 for inactive, 1 for active, -1 for"
" uninitialized");
// Now call the parent class' declareOptions
inherited::declareOptions(ol);
}

| static const PPath& PLearn::MoleculeTemplate::declaringFile | ( | ) | [inline, static] |
Reimplemented from PLearn::Molecule.
Definition at line 98 of file MoleculeTemplate.h.
:
//##### Protected Options ###############################################
| MoleculeTemplate * PLearn::MoleculeTemplate::deepCopy | ( | CopiesMap & | copies | ) | const [virtual] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| OptionList & PLearn::MoleculeTemplate::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| OptionMap & PLearn::MoleculeTemplate::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| RemoteMethodMap & PLearn::MoleculeTemplate::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Molecule.
Definition at line 57 of file MoleculeTemplate.cc.
| void PLearn::MoleculeTemplate::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Transforms a shallow copy into a deep copy.
Reimplemented from PLearn::Molecule.
Definition at line 90 of file MoleculeTemplate.cc.
References PLearn::deepCopyField(), feat_dev, and geom_dev.
{
inherited::makeDeepCopyFromShallowCopy(copies);
// deepCopyField(trainvec, copies);
deepCopyField(geom_dev, copies);
deepCopyField(feat_dev, copies);
}

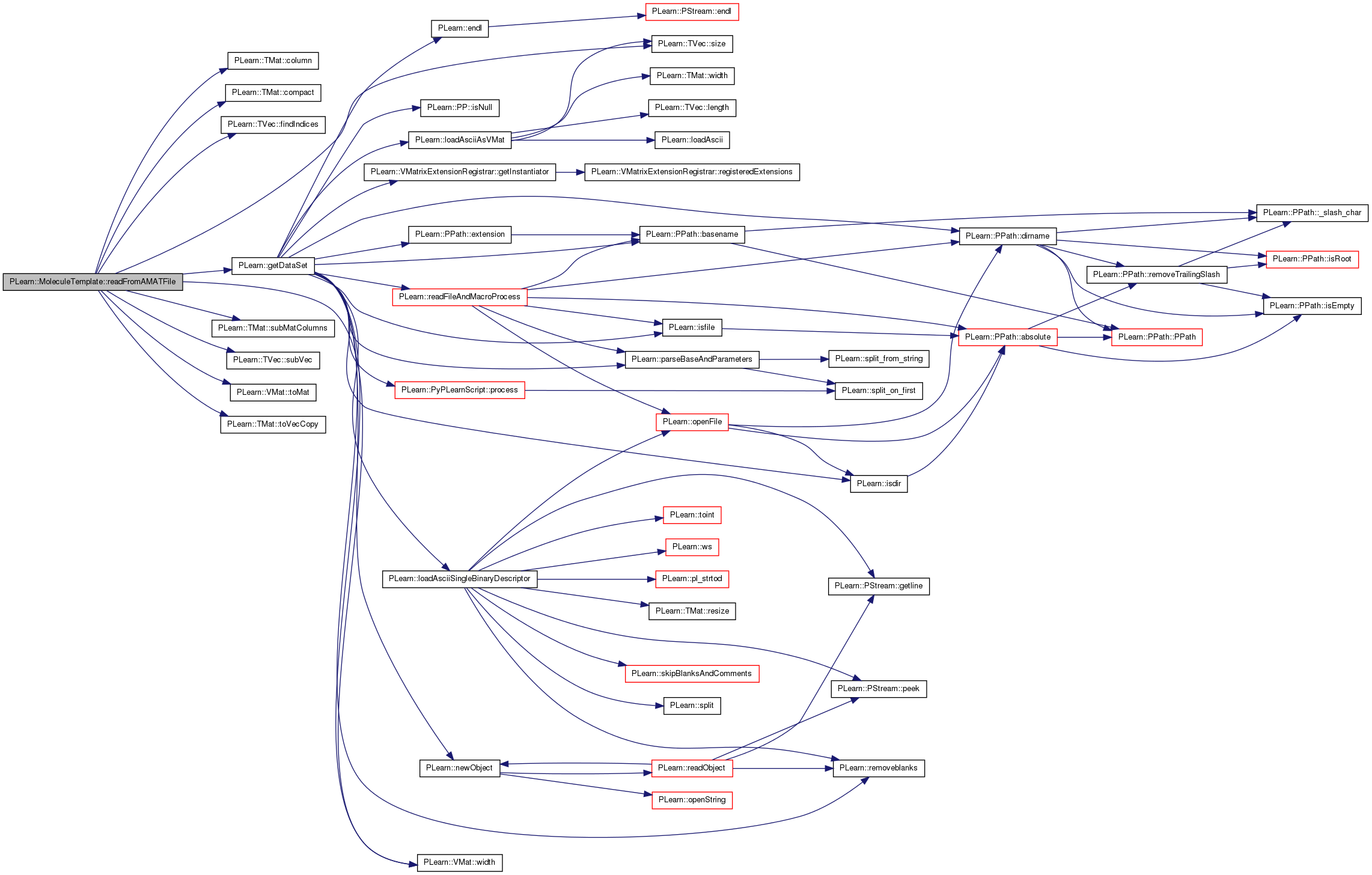
| void PLearn::MoleculeTemplate::readFromAMATFile | ( | const PPath & | filename | ) | [protected, virtual] |
Override inherited function to read also deviation info, if able.
Reimplemented from PLearn::Molecule.
Definition at line 100 of file MoleculeTemplate.cc.
References PLearn::TMat< T >::column(), PLearn::TMat< T >::compact(), feat_dev, PLearn::Molecule::feature_names, PLearn::Molecule::features, PLearn::TVec< T >::findIndices(), geom_dev, PLearn::getDataSet(), PLERROR, PLearn::TVec< T >::size(), PLearn::TMat< T >::subMatColumns(), PLearn::TVec< T >::subVec(), PLearn::VMat::toMat(), PLearn::TMat< T >::toVecCopy(), and PLearn::VMat::width().
{
VMat all = getDataSet( filename );
Mat all_mat = all->toMat();
int all_width = all->width();
TVec<string> all_names = all->fieldNames();
TVec<int> geom_dev_positions = all_names.findIndices( "geom_dev" );
if( geom_dev_positions.size() == 0 )
{
features = all_mat;
features.compact();
feature_names = all_names;
}
else if( geom_dev_positions.size() == 1 )
{
int gd_pos = geom_dev_positions[0];
if( all_width != gd_pos+1 && all_width != 2*gd_pos+1 )
PLERROR( "MoleculeTemplate::readFromAMATFile - 'geom_dev' is field"
" number %d,\n"
"there are %d following fields (expecting 0 or %d).\n",
gd_pos, all_width-gd_pos-1, gd_pos );
features = all_mat.subMatColumns(0, gd_pos);
features.compact();
geom_dev = all_mat.column( gd_pos ).toVecCopy();
feat_dev = all_mat.subMatColumns(all_width - gd_pos, gd_pos );
feat_dev.compact();
feature_names = all_names.subVec(0, gd_pos );
}
else
{
PLERROR( "MoleculeTemplate::readFromAMATFile - 'geom_dev' field should"
" be present\n"
"only once in amat file (present %d times).\n",
geom_dev_positions.size() );
}
}
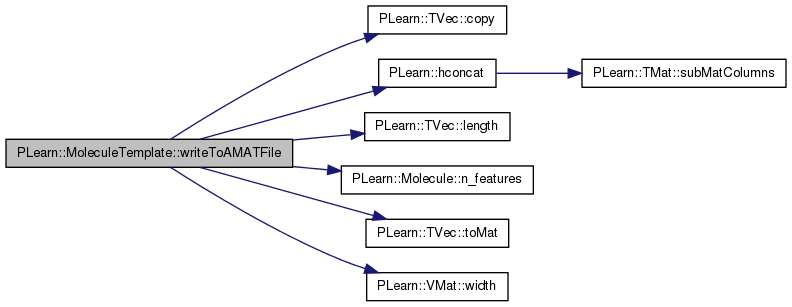
| void PLearn::MoleculeTemplate::writeToAMATFile | ( | const PPath & | filename | ) | [protected, virtual] |
Override inherited function to write also deviation info.
Reimplemented from PLearn::Molecule.
Definition at line 143 of file MoleculeTemplate.cc.
References PLearn::TVec< T >::copy(), feat_dev, PLearn::Molecule::feature_names, PLearn::Molecule::features, geom_dev, PLearn::hconcat(), i, PLearn::TVec< T >::length(), PLearn::Molecule::n_features(), PLearn::TVec< T >::toMat(), and PLearn::VMat::width().
{
VMat features_ = new MemoryVMatrix( features );
features_->declareFieldNames( feature_names );
VMat geom_dev_ = new MemoryVMatrix( geom_dev.toMat(geom_dev.length(), 1) );
TVec<string> geom_dev_names( 1, "geom_dev" );
geom_dev_->declareFieldNames( geom_dev_names );
VMat feat_dev_ = new MemoryVMatrix( feat_dev );
TVec<string> feat_dev_names = feature_names.copy();
for( int i=0 ; i<n_features() ; i++ )
feat_dev_names[i] += "_dev";
feat_dev_->declareFieldNames( feat_dev_names );
VMat all = hconcat( features_, hconcat( geom_dev_, feat_dev_ ) );
all->defineSizes( all->width(), 0, 0 );
all->saveAMAT( filename, false );
}
Reimplemented from PLearn::Molecule.
Definition at line 98 of file MoleculeTemplate.h.
class label, 0 for inactive, 1 for active, -1 for uninitialized
Definition at line 75 of file MoleculeTemplate.h.
Referenced by declareOptions().
standard deviations of each chemical property
Definition at line 72 of file MoleculeTemplate.h.
Referenced by build_(), declareOptions(), makeDeepCopyFromShallowCopy(), readFromAMATFile(), and writeToAMATFile().
### declare public option fields (such as build options) here
standard deviations of the geometrical distance
Definition at line 69 of file MoleculeTemplate.h.
Referenced by build_(), declareOptions(), makeDeepCopyFromShallowCopy(), readFromAMATFile(), and writeToAMATFile().
 1.7.4
1.7.4