|
PLearn 0.1
|
|
PLearn 0.1
|
#include <TestDependenciesCommand.h>
Public Member Functions | |
| TestDependenciesCommand () | |
| virtual void | run (const vector< string > &args) |
| The actual implementation of the 'TestDependenciesCommand' command. | |
Static Protected Attributes | |
| static PLearnCommandRegistry | reg_ |
| This allows to register the 'TestDependenciesCommand' command in the command registry. | |
Definition at line 49 of file TestDependenciesCommand.h.
| PLearn::TestDependenciesCommand::TestDependenciesCommand | ( | ) |
Definition at line 59 of file TestDependenciesCommand.cc.
: PLearnCommand("test-dependencies", "Compute dependency statistics between input and target variables.", " test-dependencies <VMat> [<inputsize> <targetsize> [<datablocksize>]]\n" "Reads a VMatrix (or any matrix format) and computes dependency statistics between each\n" "of the input variables and each of the target variables. A dependency score is then\n" "computed and a report is produced, listing the input variables in decreasing value of\n" "that score. The current implementation only computes the Spearman rank correlation\n" "and the linear correlation. If <datablocksize> is provided, it is used to\n" "divide the data row-wise in blocks of <datablocksize> rows. The statistics\n" "are computed separately in each block, and then some statistics of these\n" "statistics (min, max, mean, stdev) are reported.\n" "Missing values are ignored in the Spearman rank correlation.\n" ) {}
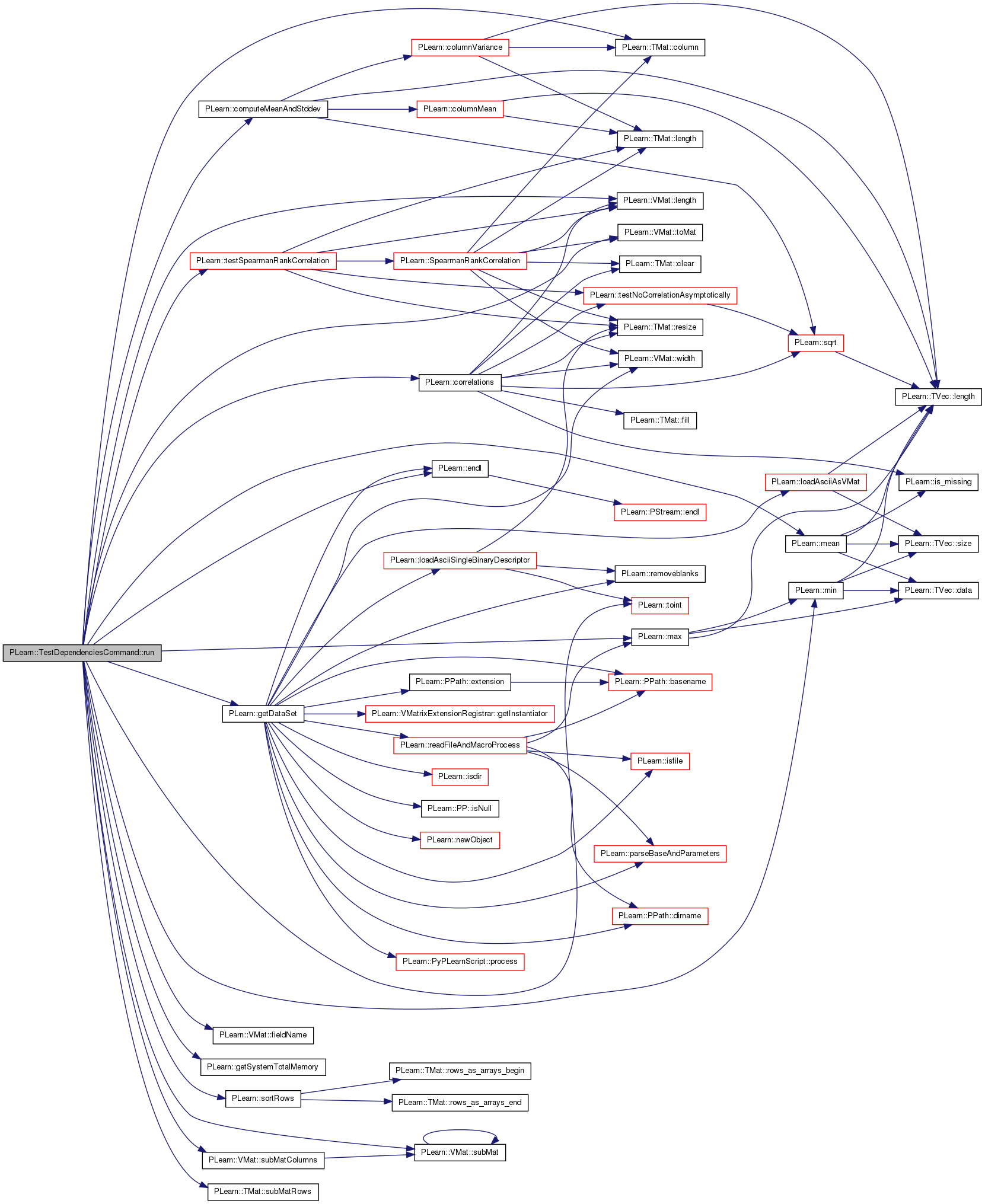
| void PLearn::TestDependenciesCommand::run | ( | const vector< string > & | args | ) | [virtual] |
The actual implementation of the 'TestDependenciesCommand' command.
Implements PLearn::PLearnCommand.
Definition at line 79 of file TestDependenciesCommand.cc.
References b, PLearn::TMat< T >::column(), PLearn::computeMeanAndStddev(), PLearn::correlations(), PLearn::endl(), PLearn::VMat::fieldName(), PLearn::getDataSet(), PLearn::getSystemTotalMemory(), i, j, PLearn::VMat::length(), PLearn::max(), PLearn::mean(), PLearn::min(), n, PLERROR, PLWARNING, PLearn::sortRows(), PLearn::VMat::subMat(), PLearn::VMat::subMatColumns(), PLearn::TMat< T >::subMatRows(), PLearn::testSpearmanRankCorrelation(), PLearn::toint(), PLearn::VMat::toMat(), and x.
{
if(args.size()<1 || args.size()>4)
PLERROR("test-dependencies expects 1 to 4 arguments, check the help");
VMat data = getDataSet(args[0]);
int inputsize = (args.size()>1)?toint(args[1]):data->inputsize();
int targetsize = (args.size()>2)?toint(args[2]):data->targetsize();
int row_blocksize = (args.size()>3)?toint(args[3]):data.length();
if (args.size()>1)
data->defineSizes(inputsize,targetsize,data->weightsize());
#ifdef WIN32
MEMORYSTATUS stat;
GlobalMemoryStatus (&stat);
// Total available memory in bytes
int memory_size = int(stat.dwAvailVirtual);
#else
int memory_size = int(getSystemTotalMemory());
#endif
int n_rowblocks = int(ceil(data.length() / real(row_blocksize)));
// statistics computed for each variable, and for each rowblock
// rank in "bestness"
// score in "bestness"
// rank correlation
// rank correlation p-value
// linear correlation
// linear correlation p-value
Mat var_rank(n_rowblocks,inputsize);
Mat var_score(n_rowblocks,inputsize);
Mat var_rank_corr(n_rowblocks,inputsize*targetsize);
Mat var_rc_pvalue(n_rowblocks,inputsize*targetsize);
Mat var_lin_corr(n_rowblocks,inputsize*targetsize);
Mat var_lc_pvalue(n_rowblocks,inputsize*targetsize);
int rowblockstart = 0;
int n=data->length();
for (int rowblock=0;rowblock<n_rowblocks;rowblock++, rowblockstart += row_blocksize)
{
int rowblocklen = (rowblock<n_rowblocks-1)?row_blocksize:(n-rowblockstart);
VMat x = data.subMat(rowblockstart,0,rowblocklen,inputsize);
VMat y = data.subMat(rowblockstart,inputsize,rowblocklen,targetsize);
Mat r = var_rank_corr(rowblock).toMat(inputsize,targetsize);
Mat pvalues = var_rc_pvalue(rowblock).toMat(inputsize,targetsize);
int col_blocksize = memory_size/int(2*sizeof(real)*rowblocklen);
if (col_blocksize>=inputsize) // everything fits in half the memory
{
x = VMat(x.toMat());
testSpearmanRankCorrelation(x,y,r,pvalues, true);
}
else // work by column blocks
{
int n_col_blocks = int(ceil(inputsize/real(col_blocksize)));
cout << "work with " << n_col_blocks << " of " << col_blocksize << " columns each (except the last)." << endl;
int bstart=0;
for (int b=0;b<n_col_blocks;b++,bstart+=col_blocksize)
{
int bsize= (b<n_col_blocks-1)?col_blocksize:inputsize-bstart;
VMat block = VMat(x.subMatColumns(bstart,bsize).toMat());
Mat rb = r.subMatRows(bstart,bsize);
Mat pb = pvalues.subMatRows(bstart,bsize);
cout << "compute rank correlation for variables " << bstart << " - " << bstart+bsize-1 << endl;
testSpearmanRankCorrelation(block,y,rb,pb, true);
}
}
// linear correlations and corresponding p-values
Mat lr = var_lin_corr(rowblock).toMat(inputsize,targetsize);
Mat lpvalues = var_lc_pvalue(rowblock).toMat(inputsize,targetsize);
correlations(x, y, lr, lpvalues, true);
Mat scores(inputsize,2);
for (int i=0;i<inputsize;i++)
{
Vec r_i = r(i);
real s =0;
for (int j=0;j<targetsize;j++)
{
real abs_r = fabs(r_i[j]);
if (abs_r>s) s=abs_r;
}
scores(i,0) = s;
scores(i,1) = i;
}
sortRows(scores,0,false);
cout << "Results for " << rowblock << "-th row block, from row " << rowblockstart << " to " << rowblockstart+rowblocklen-1 << " inclusively" << endl;
for (int k=0;k<inputsize;k++)
{
int i = int(scores(k,1));
var_rank(rowblock,i) = k;
var_score(rowblock,i) = scores(k,0);
cout << k << "-th best variable is " << data->fieldName(i) << " (col. " << i << ")";
if (targetsize==1)
cout << " with rank correlation = " << r(i,0) << " {p-value = " << pvalues(i,0)
<< "}, linear corr. = "
<< lr(i,0)
<< " {p-value= " << lpvalues(i,0) << "}" << endl;
if (targetsize>1)
{
cout << " (rank corr., rank p-value, lin. corr., lin. p-value) for individual targets: ";
for (int j=0;j<targetsize;j++)
cout << "(" << r(i,j) << ", " << pvalues(i,j) << "," << lr(i,j) << ", "
<< lpvalues(i,j) << ") ";
cout << endl;
}
}
}
// compute mean var_score for each variable and sort them accordingly
Mat mean_score(inputsize,2);
for (int i=0;i<inputsize;i++)
{
mean_score(i,0) = mean(var_score.column(i));
mean_score(i,1) = i;
}
sortRows(mean_score,0,false);
// compute statistics across row blocks
cout << "For each block statistic print (mean,stdev,min,max)\n" << endl;
for (int k=0;k<inputsize;k++)
{
int i = int(mean_score(k,1));
Mat varrank = var_rank.column(i);
Mat varscore = var_score.column(i);
Mat varrc = var_rank_corr.column(i);
Mat varrcpv = var_rc_pvalue.column(i);
Mat varlc = var_lin_corr.column(i);
Mat varlcpv = var_lc_pvalue.column(i);
Vec rankm(1),rankdev(1),scorem(1),scoredev(1),rcm(1),rcdev(1),rcpvm(1),rcpvdev(1),
lcm(1),lcdev(1),lcpvm(1),lcpvdev(1);
computeMeanAndStddev(varrank,rankm,rankdev);
computeMeanAndStddev(varscore,scorem,scoredev);
computeMeanAndStddev(varrc,rcm,rcdev);
computeMeanAndStddev(varrcpv,rcpvm,rcpvdev);
computeMeanAndStddev(varlc,lcm,lcdev);
computeMeanAndStddev(varlcpv,lcpvm,lcpvdev);
cout << k << "-th best variable is " << data->fieldName(i) << " (col. " << i << ")";
if (targetsize==1)
{
cout << " rank corr (" << rcm[0] << "," << rcdev[0] << "," << min(varrc) << "," << max(varrc) << " ) ";
cout << " var rank (" << rankm[0] << "," << rankdev[0] << "," << min(varrank) << "," << max(varrank) << " ) ";
cout << " rank cor pval(" << rcpvm[0] << "," << rcpvdev[0] << "," << min(varrcpv) << "," << max(varrcpv) << " ) ";
cout << " lin corr (" << lcm[0] << "," << lcdev[0] << "," << min(varlc) << "," << max(varlc) << " ) ";
cout << " lin cor pval (" << lcpvm[0] << "," << lcpvdev[0] << "," << min(varlcpv) << "," << max(varlcpv) << " ) " << endl;
}
else PLWARNING("In TestDependenciesCommand::run - The case 'targetsize > 1' is not implemented yet");
}
}
PLearnCommandRegistry PLearn::TestDependenciesCommand::reg_ [static, protected] |
This allows to register the 'TestDependenciesCommand' command in the command registry.
Definition at line 57 of file TestDependenciesCommand.h.
 1.7.4
1.7.4