|
PLearn 0.1
|
|
PLearn 0.1
|
#include <plearn/io/load_and_save.h>#include <plearn/math/TMat_maths.h>#include <map>#include "Molecule.h"#include "ICP.h"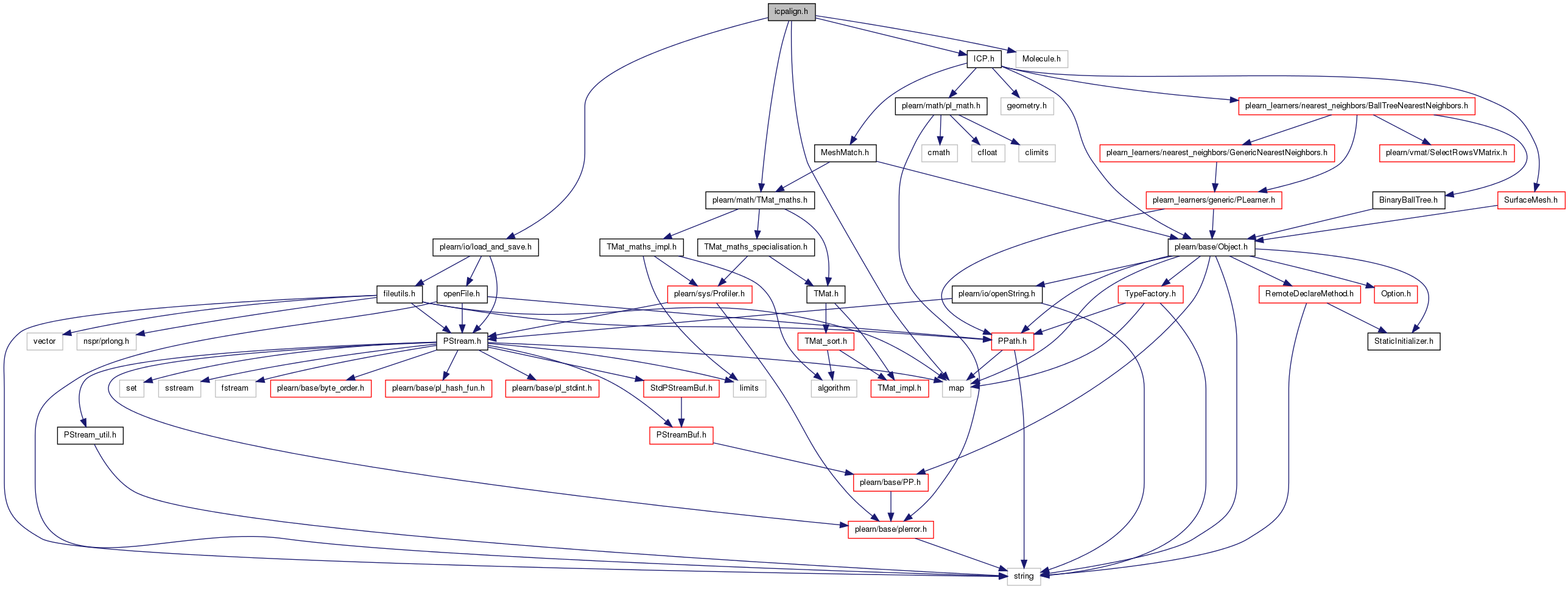
Go to the source code of this file.
Functions | |
| void | align_ (string m1_vrml_file, Mat m1_chem, string m2_vrml_file, Mat m2_chem, int rotx, int roty, int rotz, Mat &wm, double &error) |
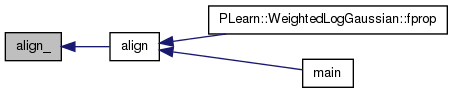
| void | align (string m1_vrml_file, Mat m1_chem, string m2_vrml_file, Mat m2_chem, Mat &wm) |
Definition at line 45 of file icpalign.h.
References align_(), PLearn::endl(), PLearn::TMat< T >::length(), and PLearn::transpose().
Referenced by PLearn::WeightedLogGaussian::fprop(), and main().
{
int m1_size = m1_chem.length();
int m2_size = m2_chem.length();
bool switched = m1_size < m2_size ;
double error = 1e10 ;
int angle = 360 ;
for(int rotx = 0 ; rotx<360 ; rotx += angle)
for(int roty = 0 ; roty<360 ; roty += angle)
for(int rotz = 0 ; rotz<360 ; rotz += angle) {
double current_error ;
Mat current_wm ;
if (switched){
align_(m2_vrml_file , m2_chem , m1_vrml_file , m1_chem , rotx,roty,rotz,current_wm,current_error) ;
}
else{
align_(m1_vrml_file , m1_chem , m2_vrml_file , m2_chem , rotx,roty,rotz,current_wm,current_error) ;
}
cout << "current error " << current_error << endl ;
if (error > current_error){
error = current_error ;
wm = current_wm ;
}
}
if (switched)
wm = transpose(wm) ;
}
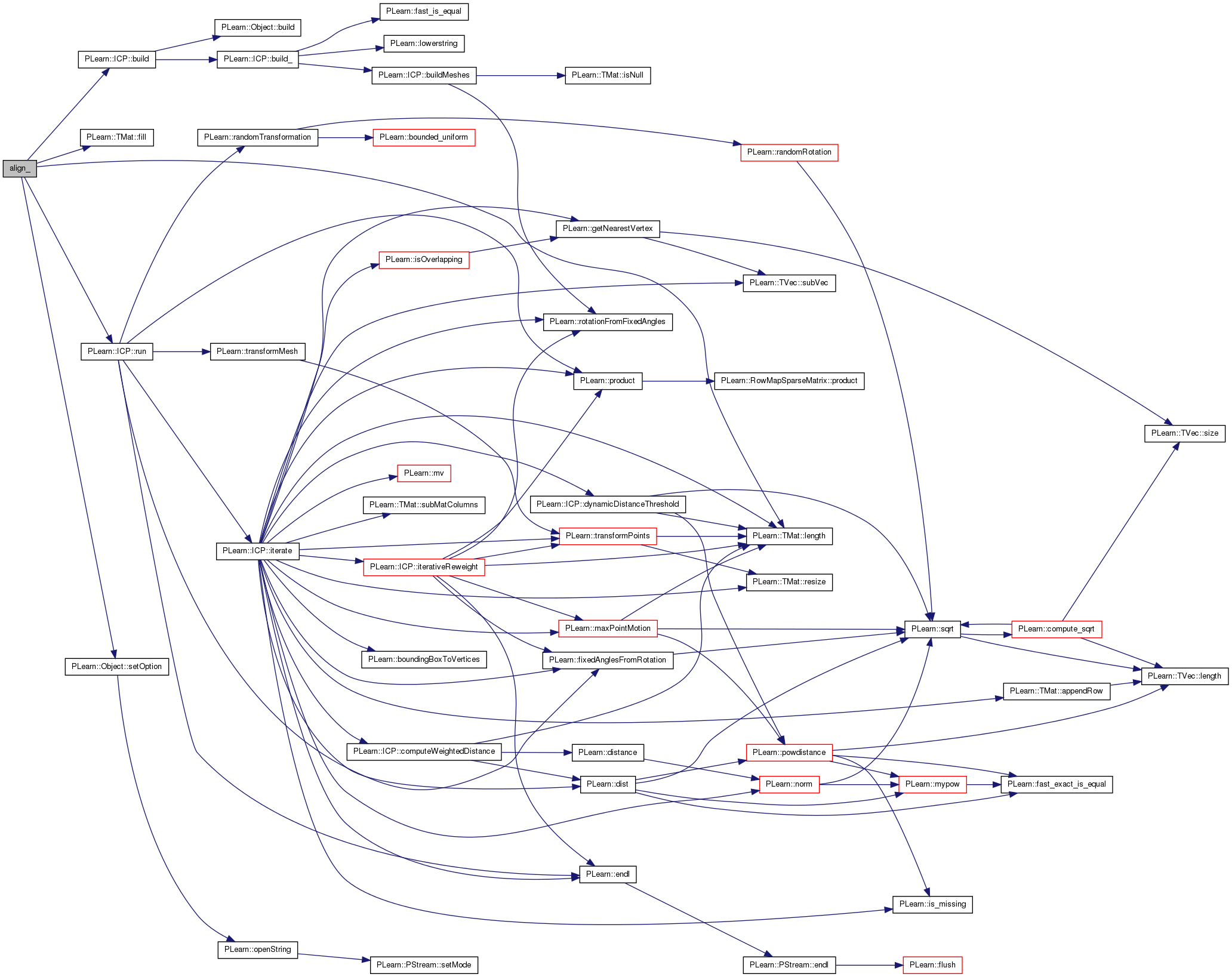
| void align_ | ( | string | m1_vrml_file, |
| Mat | m1_chem, | ||
| string | m2_vrml_file, | ||
| Mat | m2_chem, | ||
| int | rotx, | ||
| int | roty, | ||
| int | rotz, | ||
| Mat & | wm, | ||
| double & | error | ||
| ) |
Definition at line 9 of file icpalign.h.
References PLearn::ICP::build(), PLearn::TMat< T >::fill(), i, PLearn::ICP::initial_transform, PLearn::TMat< T >::length(), PLearn::ICP::match, PLearn::ICP::model_features, PLearn::ICP::run(), PLearn::ICP::scene_features, PLearn::Object::setOption(), and PLearn::ICP::vtx_matching.
Referenced by align().
{
ICP icp;
icp.model_features = m1_chem ;
icp.scene_features = m2_chem ;
icp.setOption("model_file",m1_vrml_file) ;
icp.setOption("scene_file",m2_vrml_file) ;
icp.setOption("verbosity","0") ;
icp.setOption("weight_method","oracle");
char buf[100] ;
sprintf(buf,"0 0 0 %d %d %d" , rotx, roty, rotz) ;
icp.initial_transform = string(buf) ;
icp.setOption("write_vtx_match","0");
icp.setOption("max_iter","100");
icp.setOption("error_t","0.01");
icp.setOption("dist_t","0.1");
icp.build();
icp.run();
int m1_size = m1_chem.length();
int m2_size = m2_chem.length();
wm = Mat(m1_size , m2_size) ;
wm.fill(0.0) ;
for(int i=0 ; i<icp.vtx_matching.length() ; ++i) {
wm[ (int)icp.vtx_matching[i][0] ][ (int)icp.vtx_matching[i][1] ] = 1 ;
}
error = icp.match->error ;
}
 1.7.4
1.7.4