|
PLearn 0.1
|
|
PLearn 0.1
|
Generate samples from a mixture of two gaussians. More...
#include <Preprocessing.h>
Public Member Functions | |
| Preprocessing () | |
| Default constructor. | |
| int | outputsize () const |
| SUBCLASS WRITING: override this so that it returns the size of this learner's output, as a function of its inputsize(), targetsize() and set options. | |
| void | train () |
| *** SUBCLASS WRITING: *** | |
| void | computeOutput (const Vec &, Vec &) const |
| *** SUBCLASS WRITING: *** | |
| void | computeCostsFromOutputs (const Vec &, const Vec &, const Vec &, Vec &) const |
| *** SUBCLASS WRITING: *** | |
| TVec< string > | getTestCostNames () const |
| *** SUBCLASS WRITING: *** | |
| TVec< string > | getTrainCostNames () const |
| *** SUBCLASS WRITING: *** | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual Preprocessing * | deepCopy (CopiesMap &copies) const |
| virtual void | build () |
| Finish building the object; just call inherited::build followed by build_() | |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Transforms a shallow copy into a deep copy. | |
Static Public Member Functions | |
| static string | _classname_ () |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| VMat | test_set |
| ### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //! | |
| VMat | unknown_set |
| The unknown data set. | |
| PP< ComputeDond2Target > | compute_target_learner_template |
| The template of the script to generate the class target. | |
| PP< FixDond2BinaryVariables > | fix_binary_variables_template |
| The template of the script to fix the binary variables. | |
| TVec< pair< string, string > > | imputation_spec |
| Pairs of instruction of the form field_name : mean | median | mode. | |
| TVec< pair< string, TVec< pair < real, real > > > > | discrete_variable_instructions |
| The instructions to fix the binary variables in the form of field_name : instruction. | |
| TVec< string > | selected_variables_for_input |
| The list of variables selected as input vector. | |
| TVec< string > | selected_variables_for_target |
| The list of variables selected as target vector. | |
| TVec< string > | inputs_excluded_from_gaussianization |
| The list of variables excluded from the gaussianization step. | |
| TVec< string > | targets_excluded_from_gaussianization |
| The list of variables excluded from the gaussianization step. | |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares the class options. | |
Private Types | |
| typedef PLearner | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
| void | manageTrainTestUnknownSets () |
Generate samples from a mixture of two gaussians.
Definition at line 60 of file Preprocessing.h.
typedef PLearner PLearn::Preprocessing::inherited [private] |
Reimplemented from PLearn::PLearner.
Definition at line 62 of file Preprocessing.h.
| PLearn::Preprocessing::Preprocessing | ( | ) |
| string PLearn::Preprocessing::_classname_ | ( | ) | [static] |
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
| OptionList & PLearn::Preprocessing::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
| RemoteMethodMap & PLearn::Preprocessing::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
| Object * PLearn::Preprocessing::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Definition at line 57 of file Preprocessing.cc.
| StaticInitializer Preprocessing::_static_initializer_ & PLearn::Preprocessing::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
| void PLearn::Preprocessing::build | ( | ) | [virtual] |
Finish building the object; just call inherited::build followed by build_()
Reimplemented from PLearn::PLearner.
Definition at line 129 of file Preprocessing.cc.
{
// ### Nothing to add here, simply calls build_().
inherited::build();
build_();
}
| void PLearn::Preprocessing::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::PLearner.
Definition at line 139 of file Preprocessing.cc.
References PLearn::endl(), and PLERROR.
{
MODULE_LOG << "build_() called" << endl;
if (train_set)
{
manageTrainTestUnknownSets();
PLERROR("In Preprocessing: Everything completed successfuly, we are done here");
}
}

| string PLearn::Preprocessing::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 57 of file Preprocessing.cc.
| void PLearn::Preprocessing::computeCostsFromOutputs | ( | const Vec & | input, |
| const Vec & | output, | ||
| const Vec & | target, | ||
| Vec & | costs | ||
| ) | const [virtual] |
*** SUBCLASS WRITING: ***
This should be defined in subclasses to compute the weighted costs from already computed output. The costs should correspond to the cost names returned by getTestCostNames().
NOTE: In exotic cases, the cost may also depend on some info in the input, that's why the method also gets so see it.
Implements PLearn::PLearner.
Definition at line 961 of file Preprocessing.cc.
{}
*** SUBCLASS WRITING: ***
This should be defined in subclasses to compute the output from the input.
Reimplemented from PLearn::PLearner.
Definition at line 960 of file Preprocessing.cc.
{}
| void PLearn::Preprocessing::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares the class options.
Reimplemented from PLearn::PLearner.
Definition at line 69 of file Preprocessing.cc.
References PLearn::OptionBase::buildoption, compute_target_learner_template, PLearn::declareOption(), discrete_variable_instructions, fix_binary_variables_template, imputation_spec, inputs_excluded_from_gaussianization, selected_variables_for_input, selected_variables_for_target, targets_excluded_from_gaussianization, test_set, and unknown_set.
{
declareOption(ol, "test_set", &Preprocessing::test_set,
OptionBase::buildoption,
"The test data set.\n");
declareOption(ol, "unknown_set", &Preprocessing::unknown_set,
OptionBase::buildoption,
"The unknown data set.\n");
declareOption(ol, "compute_target_learner_template", &Preprocessing::compute_target_learner_template,
OptionBase::buildoption,
"The template of the script to generate the class target.\n");
declareOption(ol, "fix_binary_variables_template", &Preprocessing::fix_binary_variables_template,
OptionBase::buildoption,
"The template of the script to fix the binary variables.\n");
declareOption(ol, "imputation_spec", &Preprocessing::imputation_spec,
OptionBase::buildoption,
"Pairs of instruction of the form field_name : mean | median | mode.\n");
declareOption(ol, "discrete_variable_instructions", &Preprocessing::discrete_variable_instructions,
OptionBase::buildoption,
"The instructions to dichotomize the variables in the form of field_name : TVec<pair>.\n"
"The pairs are values from : to, each creating a 0, 1 variable.\n"
"Variables with no specification will be kept as_is.\n");
declareOption(ol, "selected_variables_for_input", &Preprocessing::selected_variables_for_input,
OptionBase::buildoption,
"The list of variables selected as input vector.\n");
declareOption(ol, "selected_variables_for_target", &Preprocessing::selected_variables_for_target,
OptionBase::buildoption,
"The list of variables selected as target vector.\n");
declareOption(ol, "inputs_excluded_from_gaussianization", &Preprocessing::inputs_excluded_from_gaussianization,
OptionBase::buildoption,
"The list of input variables excluded from the gaussianization step.\n");
declareOption(ol, "targets_excluded_from_gaussianization", &Preprocessing::targets_excluded_from_gaussianization,
OptionBase::buildoption,
"The list of target variables excluded from the gaussianization step.\n");
inherited::declareOptions(ol);
}

| static const PPath& PLearn::Preprocessing::declaringFile | ( | ) | [inline, static] |
Reimplemented from PLearn::PLearner.
Definition at line 114 of file Preprocessing.h.
:
//##### Protected Member Functions ######################################
| Preprocessing * PLearn::Preprocessing::deepCopy | ( | CopiesMap & | copies | ) | const [virtual] |
Reimplemented from PLearn::PLearner.
Definition at line 57 of file Preprocessing.cc.
| OptionList & PLearn::Preprocessing::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 57 of file Preprocessing.cc.
| OptionMap & PLearn::Preprocessing::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 57 of file Preprocessing.cc.
| RemoteMethodMap & PLearn::Preprocessing::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 57 of file Preprocessing.cc.
| TVec< string > PLearn::Preprocessing::getTestCostNames | ( | ) | const [virtual] |
*** SUBCLASS WRITING: ***
This should return the names of the costs computed by computeCostsFromOutputs.
Implements PLearn::PLearner.
Definition at line 962 of file Preprocessing.cc.
References PLearn::TVec< T >::append().
{
TVec<string> result;
result.append( "MSE" );
return result;
}

| TVec< string > PLearn::Preprocessing::getTrainCostNames | ( | ) | const [virtual] |
*** SUBCLASS WRITING: ***
This should return the names of the objective costs that the train method computes and for which it updates the VecStatsCollector train_stats.
Implements PLearn::PLearner.
Definition at line 968 of file Preprocessing.cc.
References PLearn::TVec< T >::append().
{
TVec<string> result;
result.append( "MSE" );
return result;
}

| void PLearn::Preprocessing::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Transforms a shallow copy into a deep copy.
Reimplemented from PLearn::PLearner.
Definition at line 110 of file Preprocessing.cc.
References PLearn::deepCopyField().
{
deepCopyField(test_set, copies);
deepCopyField(unknown_set, copies);
deepCopyField(compute_target_learner_template, copies);
deepCopyField(fix_binary_variables_template, copies);
deepCopyField(imputation_spec, copies);
deepCopyField(discrete_variable_instructions, copies);
deepCopyField(selected_variables_for_input, copies);
deepCopyField(selected_variables_for_target, copies);
deepCopyField(inputs_excluded_from_gaussianization, copies);
deepCopyField(targets_excluded_from_gaussianization, copies);
inherited::makeDeepCopyFromShallowCopy(copies);
}

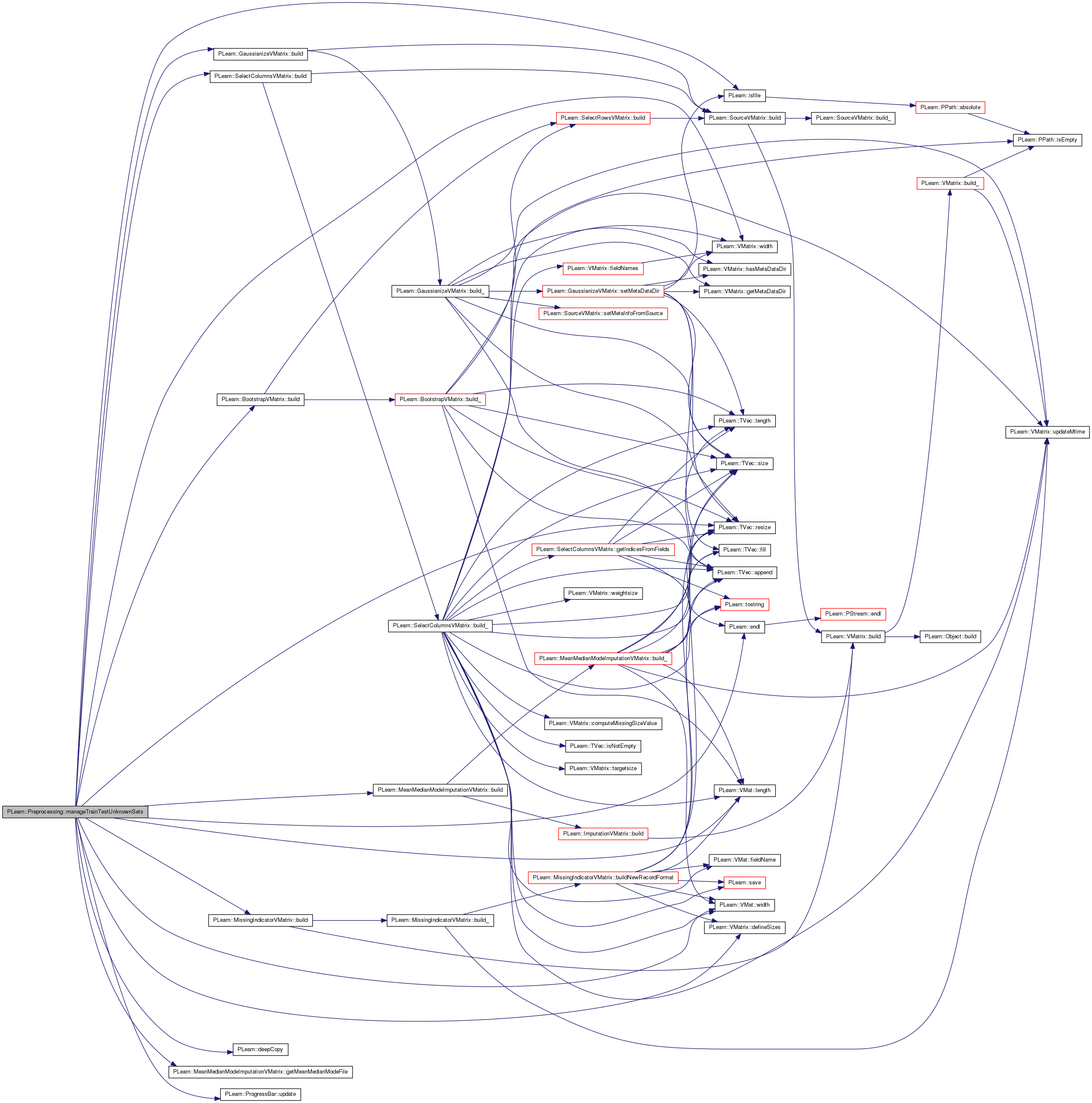
| void PLearn::Preprocessing::manageTrainTestUnknownSets | ( | ) | [private] |
Definition at line 149 of file Preprocessing.cc.
References PLearn::GaussianizeVMatrix::build(), PLearn::MissingIndicatorVMatrix::build(), PLearn::SelectColumnsVMatrix::build(), PLearn::MeanMedianModeImputationVMatrix::build(), PLearn::BootstrapVMatrix::build(), PLearn::deepCopy(), PLearn::VMatrix::defineSizes(), PLearn::endl(), PLearn::SelectColumnsVMatrix::extend_with_missing, PLearn::SelectColumnsVMatrix::fields, PLearn::SelectColumnsVMatrix::fields_partial_match, PLearn::BootstrapVMatrix::frac, PLearn::GaussianizeVMatrix::gaussianize_extra, PLearn::GaussianizeVMatrix::gaussianize_input, PLearn::GaussianizeVMatrix::gaussianize_target, PLearn::GaussianizeVMatrix::gaussianize_weight, PLearn::MeanMedianModeImputationVMatrix::getMeanMedianModeFile(), PLearn::MeanMedianModeImputationVMatrix::imputation_spec, PLearn::isfile(), PLearn::VMat::length(), PLearn::MissingIndicatorVMatrix::number_of_train_samples_to_use, PLearn::MeanMedianModeImputationVMatrix::number_of_train_samples_to_use, PLearn::BootstrapVMatrix::own_seed, PLearn::TVec< T >::resize(), PLearn::BootstrapVMatrix::shuffle, PLearn::MissingIndicatorVMatrix::source, PLearn::ImputationVMatrix::source, PLearn::SourceVMatrix::source, PLearn::GaussianizeVMatrix::threshold_ratio, PLearn::MeanMedianModeImputationVMatrix::train_set, PLearn::MissingIndicatorVMatrix::train_set, PLearn::GaussianizeVMatrix::train_source, PLearn::ProgressBar::update(), PLearn::VMat::width(), and PLearn::VMatrix::width().
{
// defining all the variables for the train set
PPath output_path;
PPath train_with_class_target_file_name;
VMat train_with_class_target_file;
PP<ComputeDond2Target> compute_target_learner;
VMat train_shuffled_file;
PPath train_with_binary_fixed_file_name;
VMat train_with_binary_fixed_file;
PP<FixDond2BinaryVariables> fix_binary_variables_learner;
PPath train_with_ind_file_name;
VMat train_with_ind_vmat;
VMat train_with_ind_file;
Vec train_with_ind_vector;
VMat mean_median_mode_with_ind_file;
PPath train_with_dichotomies_file_name;
VMat train_with_dichotomies_file;
PPath mean_median_mode_with_dichotmies_file_name;
VMat mean_median_mode_with_dichotmies_file;
PP<DichotomizeDond2DiscreteVariables> dichotomize_discrete_variables_learner;
SelectColumnsVMatrix* train_with_selected_columns_vmatrix;
VMat train_with_selected_columns_vmat;
VMat mean_median_mode_with_selected_columns_vmat;
VMat train_gaussianized_vmat;
GaussianizeVMatrix* mean_median_mode_gaussianized_vmatrix;
VMat mean_median_mode_gaussianized_vmat;
PPath train_input_preprocessed_file_name;
VMat train_input_preprocessed_file;
Vec train_input_preprocessed_vector;
PPath mean_median_mode_input_preprocessed_file_name;
VMat mean_median_mode_input_preprocessed_file;
Vec mean_median_mode_input_preprocessed_vector;
SelectColumnsVMatrix* train_target_with_selected_columns_vmatrix;
VMat train_target_with_selected_columns_vmat;
GaussianizeVMatrix* train_target_gaussianized_vmatrix;
VMat train_target_gaussianized_vmat;
PPath train_target_preprocessed_file_name;
VMat train_target_preprocessed_file;
Vec train_target_preprocessed_vector;
ProgressBar* pb = 0;
// managing the train set
cout << "In Preprocessing: we start by formatting the training set" << endl;
cout << endl << "****** STEP 1 ******" << endl;
cout << "The first step groups variables by type, skips untrustworthy variables, and generate class targets" << endl;
cout << "It uses ComputeDond2Target to transform base_train.pmat into step1_train_with_class_target.pmat" << endl;
output_path = expdir+"step1_train_with_class_target";
cout << "output_path" << output_path;
train_with_class_target_file_name = output_path + ".pmat";
if (isfile(train_with_class_target_file_name))
{
train_with_class_target_file = new FileVMatrix(train_with_class_target_file_name);
train_with_class_target_file->defineSizes(train_with_class_target_file->width(), 0, 0);
cout << train_with_class_target_file_name << " already exist, we are skipping this step." << endl;
}
else
{
compute_target_learner = ::PLearn::deepCopy(compute_target_learner_template);
compute_target_learner->unknown_sales = 0;
compute_target_learner->output_path = output_path;
compute_target_learner->setTrainingSet(train_set, true);
train_with_class_target_file = compute_target_learner->getOutputFile();
}
cout << endl << "****** STEP 2 ******" << endl;
cout << "This step shuffles the training set to get training data in random order." << endl;
cout << "It uses BootstrapVMatrix to transform step1_train_with_class_target.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 3" << endl;
output_path = expdir+"step3_train_with_binary_fixed";
train_with_binary_fixed_file_name = output_path + ".pmat";
if (isfile(train_with_binary_fixed_file_name))
{
cout << train_with_binary_fixed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
BootstrapVMatrix* train_shufffled_vmatrix = new BootstrapVMatrix();
train_shufffled_vmatrix->shuffle = 1;
train_shufffled_vmatrix->frac = 1.0;
train_shufffled_vmatrix->own_seed = 123456;
train_shufffled_vmatrix->source = train_with_class_target_file;
train_shufffled_vmatrix->build();
train_shuffled_file = train_shufffled_vmatrix;
}
cout << endl << "****** STEP 3 ******" << endl;
cout << "For strictly binary variables, various situations arise: zero or non-zero, missing or not-missing, a given value or not, etc..." << endl;
cout << "This step uses FixDond2BinaryVariables to create step3_train_with_binary_fixed.pmat with 0-1 binary variables." << endl;
if (isfile(train_with_binary_fixed_file_name))
{
train_with_binary_fixed_file = new FileVMatrix(train_with_binary_fixed_file_name);
train_with_binary_fixed_file->defineSizes(train_with_binary_fixed_file->width(), 0, 0);
cout << train_with_binary_fixed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
fix_binary_variables_learner = ::PLearn::deepCopy(fix_binary_variables_template);
fix_binary_variables_learner->output_path = output_path;
fix_binary_variables_learner->setTrainingSet(train_shuffled_file, true);
train_with_binary_fixed_file = fix_binary_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 4 ******" << endl;
cout << "This step adds missing indicators variables to each variable with missing values." << endl;
cout << "It uses MissingIndicatorVMatrix to transform step3_train_with_binary_fixed.pmat" << endl;
cout << "The resulting vitual view is stored in step4_train_with_ind.pmat." << endl;
train_with_ind_file_name = "step4_train_with_ind.pmat";
if (isfile(train_with_ind_file_name))
{
train_with_ind_file = new FileVMatrix(train_with_ind_file_name);
train_with_ind_file->defineSizes(train_with_ind_file->width(), 0, 0);
cout << train_with_ind_file_name << " already exist, we are skipping this step." << endl;
}
else
{
MissingIndicatorVMatrix* train_with_ind_vmatrix = new MissingIndicatorVMatrix();
train_with_ind_vmatrix->source = train_with_binary_fixed_file;
train_with_ind_vmatrix->train_set = train_with_binary_fixed_file;
train_with_ind_vmatrix->number_of_train_samples_to_use = 30000.0;
train_with_ind_vmatrix->build();
train_with_ind_vmat = train_with_ind_vmatrix;
train_with_ind_file = new FileVMatrix(train_with_ind_file_name, train_with_ind_vmat->length(), train_with_ind_vmat->fieldNames());
train_with_ind_file->defineSizes(train_with_ind_vmat->inputsize(), train_with_ind_vmat->targetsize(), train_with_ind_vmat->weightsize());
pb = new ProgressBar("Saving the train file with missing indicators", train_with_ind_vmat->length());
train_with_ind_vector.resize(train_with_ind_vmat->width());
for (int train_with_ind_row = 0; train_with_ind_row < train_with_ind_vmat->length(); train_with_ind_row++)
{
train_with_ind_vmat->getRow(train_with_ind_row, train_with_ind_vector);
train_with_ind_file->putRow(train_with_ind_row, train_with_ind_vector);
pb->update( train_with_ind_row );
}
delete pb;
}
cout << endl << "****** STEP 5 ******" << endl;
cout << "This step computes the mean, median and mode vectors on step4_train_with_ind.pmat." << endl;
cout << "The vectors are kept in the mean_median_mode_file.pmat of the metadata." << endl;
cout << "It uses MeanMedianModeImputationVMatrix to do that" << endl;
cout << "The resulting vitual view is not used." << endl;
cout << "But the mean, median and mode vectors have to go thru the same transformation than the training file" << endl;
cout << "from here on to the end of the preprocessing steps.." << endl;
{
MeanMedianModeImputationVMatrix* train_with_imp_vmatrix = new MeanMedianModeImputationVMatrix();
train_with_imp_vmatrix->source = train_with_ind_file;
train_with_imp_vmatrix->train_set = train_with_ind_file;
train_with_imp_vmatrix->number_of_train_samples_to_use = 30000.0;
train_with_imp_vmatrix->imputation_spec = imputation_spec;
train_with_imp_vmatrix->build();
mean_median_mode_with_ind_file = train_with_imp_vmatrix->getMeanMedianModeFile();
}
cout << endl << "****** STEP 6 ******" << endl;
cout << "This steps generates as many dichotomized variables as there are significant code values." << endl;
cout << "It uses DichotomizeDond2DiscreteVariables to transform step4_train_with_ind.pmat into step6_train_with_dichotomies.pmat" << endl;
output_path = expdir+"step6_train_with_dichotomies";
train_with_dichotomies_file_name = output_path + ".pmat";
if (isfile(train_with_dichotomies_file_name))
{
train_with_dichotomies_file = new FileVMatrix(train_with_dichotomies_file_name);
train_with_dichotomies_file->defineSizes(train_with_dichotomies_file->width(), 0, 0);
cout << train_with_dichotomies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
dichotomize_discrete_variables_learner = new DichotomizeDond2DiscreteVariables();
dichotomize_discrete_variables_learner->discrete_variable_instructions = discrete_variable_instructions;
dichotomize_discrete_variables_learner->output_path = output_path;
dichotomize_discrete_variables_learner->setTrainingSet(train_with_ind_file, true);
train_with_dichotomies_file = dichotomize_discrete_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 7 ******" << endl;
cout << "This steps does the same thing to the mean, median and mode vectors." << endl;
cout << "It uses DichotomizeDond2DiscreteVariables to transform step4_train_with_ind.pmat.metadata/mean_median_mode_file.pmat "
<< "into step6_train_with_dichotomies.pmat.metadata/mean_median_mode_file.pmat" << endl;
output_path = expdir+train_with_dichotomies_file_name + ".metadata/mean_median_mode_file";
mean_median_mode_with_dichotmies_file_name = output_path + ".pmat";
if (isfile(mean_median_mode_with_dichotmies_file_name))
{
mean_median_mode_with_dichotmies_file = new FileVMatrix(mean_median_mode_with_dichotmies_file_name);
mean_median_mode_with_dichotmies_file->defineSizes(mean_median_mode_with_dichotmies_file->width(), 0, 0);
cout << mean_median_mode_with_dichotmies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
dichotomize_discrete_variables_learner = new DichotomizeDond2DiscreteVariables();
dichotomize_discrete_variables_learner->discrete_variable_instructions = discrete_variable_instructions;
dichotomize_discrete_variables_learner->output_path = output_path;
dichotomize_discrete_variables_learner->setTrainingSet(mean_median_mode_with_ind_file, true);
mean_median_mode_with_dichotmies_file = dichotomize_discrete_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 8 ******" << endl;
cout << "This step select the desired columns from the training set to create the input records." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_train_with_dichotomies.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 10" << endl;
output_path = expdir+"final_train_input_preprocessed";
train_input_preprocessed_file_name = output_path + ".pmat";
if (isfile(train_input_preprocessed_file_name))
{
cout << train_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
train_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
train_with_selected_columns_vmatrix->source = train_with_dichotomies_file;
train_with_selected_columns_vmatrix->fields_partial_match = 0;
train_with_selected_columns_vmatrix->extend_with_missing = 0;
train_with_selected_columns_vmatrix->fields = selected_variables_for_input;
train_with_selected_columns_vmatrix->build();
train_with_selected_columns_vmatrix->defineSizes(train_with_selected_columns_vmatrix->width(), 0, 0);
train_with_selected_columns_vmat = train_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 9 ******" << endl;
cout << "This step does the same thing to the mean, median and mode vectors." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_train_with_dichotomies.pmat.metadata/mean_median_mode_file.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 11" << endl;
output_path = expdir+train_input_preprocessed_file_name + ".metadata/mean_median_mode_file";
mean_median_mode_input_preprocessed_file_name = output_path + ".pmat";
if (isfile(mean_median_mode_input_preprocessed_file_name))
{
cout << mean_median_mode_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
SelectColumnsVMatrix* mean_median_mode_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
mean_median_mode_with_selected_columns_vmatrix->source = mean_median_mode_with_dichotmies_file;
mean_median_mode_with_selected_columns_vmatrix->fields_partial_match = 0;
mean_median_mode_with_selected_columns_vmatrix->extend_with_missing = 0;
mean_median_mode_with_selected_columns_vmatrix->fields = selected_variables_for_input;
mean_median_mode_with_selected_columns_vmatrix->build();
mean_median_mode_with_selected_columns_vmatrix->defineSizes(mean_median_mode_with_selected_columns_vmatrix->width(), 0, 0);
mean_median_mode_with_selected_columns_vmat = mean_median_mode_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 10 ******" << endl;
cout << "This gaussianizes the input records." << endl;
cout << "It uses GaussianizeVMatrix to transform the vmat from step 8" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 12" << endl;
if (isfile(train_input_preprocessed_file_name))
{
cout << train_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
GaussianizeVMatrix* train_gaussianized_vmatrix = new GaussianizeVMatrix();
train_gaussianized_vmatrix->source = train_with_selected_columns_vmat;
train_gaussianized_vmatrix->train_source = train_with_selected_columns_vmat;
train_gaussianized_vmatrix->threshold_ratio = 1;
train_gaussianized_vmatrix->gaussianize_input = 1;
train_gaussianized_vmatrix->gaussianize_target = 0;
train_gaussianized_vmatrix->gaussianize_weight = 0;
train_gaussianized_vmatrix->gaussianize_extra = 0;
train_gaussianized_vmatrix->excluded_fields = inputs_excluded_from_gaussianization;
train_gaussianized_vmatrix->build();
train_gaussianized_vmat = train_gaussianized_vmatrix;
}
cout << endl << "****** STEP 11 ******" << endl;
cout << "This step does the same thing to the mean, median and mode vectors." << endl;
cout << "It uses the vmat from step 9" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 13" << endl;
if (isfile(mean_median_mode_input_preprocessed_file_name))
{
cout << mean_median_mode_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
mean_median_mode_gaussianized_vmatrix = new GaussianizeVMatrix();
mean_median_mode_gaussianized_vmatrix->source = mean_median_mode_with_selected_columns_vmat;
mean_median_mode_gaussianized_vmatrix->train_source = train_with_selected_columns_vmat;
mean_median_mode_gaussianized_vmatrix->threshold_ratio = 1;
mean_median_mode_gaussianized_vmatrix->gaussianize_input = 1;
mean_median_mode_gaussianized_vmatrix->gaussianize_target = 0;
mean_median_mode_gaussianized_vmatrix->gaussianize_weight = 0;
mean_median_mode_gaussianized_vmatrix->gaussianize_extra = 0;
mean_median_mode_gaussianized_vmatrix->excluded_fields = inputs_excluded_from_gaussianization;
mean_median_mode_gaussianized_vmatrix->build();
mean_median_mode_gaussianized_vmat = mean_median_mode_gaussianized_vmatrix;
}
cout << endl << "****** STEP 12 ******" << endl;
cout << "Finaly, the preprocessed input vectors are store on disk." << endl;
cout << "The vmat from step 10 is converted to final_train_input_preprocessed.pmat." << endl;
if (isfile(train_input_preprocessed_file_name))
{
cout << train_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
train_input_preprocessed_file = new FileVMatrix(train_input_preprocessed_file_name, train_gaussianized_vmat->length(), train_gaussianized_vmat->fieldNames());
train_input_preprocessed_file->defineSizes(train_gaussianized_vmat->inputsize(), train_gaussianized_vmat->targetsize(), train_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final train preprocessed input records", train_gaussianized_vmat->length());
train_input_preprocessed_vector.resize(train_gaussianized_vmat->width());
for (int train_gaussianized_row = 0; train_gaussianized_row < train_gaussianized_vmat->length(); train_gaussianized_row++)
{
train_gaussianized_vmat->getRow(train_gaussianized_row, train_input_preprocessed_vector);
train_input_preprocessed_file->putRow(train_gaussianized_row, train_input_preprocessed_vector);
pb->update( train_gaussianized_row );
}
delete pb;
}
cout << endl << "****** STEP 13 ******" << endl;
cout << "And we do the same for the mean, median and mode vectors." << endl;
cout << "The vmat from step 11 is converted to final_train_input_preprocessed.pmat.metadata/men_median_mode_file.pmat" << endl;
if (isfile(mean_median_mode_input_preprocessed_file_name))
{
cout << mean_median_mode_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
mean_median_mode_input_preprocessed_file =
new FileVMatrix(mean_median_mode_input_preprocessed_file_name, mean_median_mode_gaussianized_vmat->length(), mean_median_mode_gaussianized_vmat->fieldNames());
mean_median_mode_input_preprocessed_file->defineSizes(mean_median_mode_gaussianized_vmat->inputsize(),
mean_median_mode_gaussianized_vmat->targetsize(), mean_median_mode_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final mean,median and mode preprocessed input vectors", mean_median_mode_gaussianized_vmat->length());
mean_median_mode_input_preprocessed_vector.resize(mean_median_mode_gaussianized_vmat->width());
for (int mean_median_mode_gaussianized_row = 0; mean_median_mode_gaussianized_row < mean_median_mode_gaussianized_vmat->length(); mean_median_mode_gaussianized_row++)
{
mean_median_mode_gaussianized_vmat->getRow(mean_median_mode_gaussianized_row, mean_median_mode_input_preprocessed_vector);
mean_median_mode_input_preprocessed_file->putRow(mean_median_mode_gaussianized_row, mean_median_mode_input_preprocessed_vector);
pb->update( mean_median_mode_gaussianized_row );
}
delete pb;
}
cout << endl << "****** STEP 14 ******" << endl;
cout << "This step select the desired columns from the training set to create the target records." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_train_with_dichotomies.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 15" << endl;
output_path = expdir+"final_train_target_preprocessed";
train_target_preprocessed_file_name = output_path + ".pmat";
if (isfile(train_target_preprocessed_file_name))
{
cout << train_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
train_target_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
train_target_with_selected_columns_vmatrix->source = train_with_dichotomies_file;
train_target_with_selected_columns_vmatrix->fields_partial_match = 0;
train_target_with_selected_columns_vmatrix->extend_with_missing = 0;
train_target_with_selected_columns_vmatrix->fields = selected_variables_for_target;
train_target_with_selected_columns_vmatrix->build();
train_target_with_selected_columns_vmatrix->defineSizes(train_target_with_selected_columns_vmatrix->width(), 0, 0);
train_target_with_selected_columns_vmat = train_target_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 15 ******" << endl;
cout << "This gaussianizes the input records." << endl;
cout << "It uses GaussianizeVMatrix to transform the vmat from step 14" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 16" << endl;
if (isfile(train_target_preprocessed_file_name))
{
cout << train_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
train_target_gaussianized_vmatrix = new GaussianizeVMatrix();
train_target_gaussianized_vmatrix->source = train_target_with_selected_columns_vmat;
train_target_gaussianized_vmatrix->train_source = train_target_with_selected_columns_vmat;
train_target_gaussianized_vmatrix->threshold_ratio = 1;
train_target_gaussianized_vmatrix->gaussianize_input = 1;
train_target_gaussianized_vmatrix->gaussianize_target = 0;
train_target_gaussianized_vmatrix->gaussianize_weight = 0;
train_target_gaussianized_vmatrix->gaussianize_extra = 0;
train_target_gaussianized_vmatrix->excluded_fields = targets_excluded_from_gaussianization;;
train_target_gaussianized_vmatrix->build();
train_target_gaussianized_vmat = train_target_gaussianized_vmatrix;
}
cout << endl << "****** STEP 16 ******" << endl;
cout << "Finaly, the preprocessed input vectors are store on disk." << endl;
cout << "The vmat from step 15 is converted to final_train_target_preprocessed.pmat." << endl;
if (isfile(train_target_preprocessed_file_name))
{
cout << train_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
train_target_preprocessed_file = new FileVMatrix(train_target_preprocessed_file_name, train_target_gaussianized_vmat->length(),
train_target_gaussianized_vmat->fieldNames());
train_target_preprocessed_file->defineSizes(train_target_gaussianized_vmat->inputsize(), train_target_gaussianized_vmat->targetsize(),
train_target_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final train preprocessed target records", train_target_gaussianized_vmat->length());
train_target_preprocessed_vector.resize(train_target_gaussianized_vmat->width());
for (int train_gaussianized_row = 0; train_gaussianized_row < train_target_gaussianized_vmat->length(); train_gaussianized_row++)
{
train_target_gaussianized_vmat->getRow(train_gaussianized_row, train_target_preprocessed_vector);
train_target_preprocessed_file->putRow(train_gaussianized_row, train_target_preprocessed_vector);
pb->update( train_gaussianized_row );
}
delete pb;
}
// defining all the variables for the test set
PPath test_with_class_target_file_name;
VMat test_with_class_target_file;
PPath test_with_binary_fixed_file_name;
VMat test_with_binary_fixed_file;
PPath test_with_ind_file_name;
MissingIndicatorVMatrix* test_with_ind_vmatrix;
VMat test_with_ind_vmat;
PPath test_with_dichotomies_file_name;
VMat test_with_dichotomies_file;
SelectColumnsVMatrix* test_with_selected_columns_vmatrix;
VMat test_with_selected_columns_vmat;
GaussianizeVMatrix* test_gaussianized_vmatrix;
VMat test_gaussianized_vmat;
PPath test_input_preprocessed_file_name;
VMat test_input_preprocessed_file;
Vec test_input_preprocessed_vector;
SelectColumnsVMatrix* test_target_with_selected_columns_vmatrix;
VMat test_target_with_selected_columns_vmat;
GaussianizeVMatrix* test_target_gaussianized_vmatrix;
VMat test_target_gaussianized_vmat;
PPath test_target_preprocessed_file_name;
VMat test_target_preprocessed_file;
Vec test_target_preprocessed_vector;
// managing the test set
cout << endl << "********************" << endl;
cout << "In Preprocessing: now, we format the test set" << endl;
cout << endl << "****** STEP 1 ******" << endl;
cout << "The first step groups variables by type, skips untrustworthy variables, and generate class targets" << endl;
cout << "It uses ComputeDond2Target to transform base_test.pmat into step1_test_with_class_target.pmat" << endl;
output_path = expdir+"step1_test_with_class_target";
test_with_class_target_file_name = output_path + ".pmat";
if (isfile(test_with_class_target_file_name))
{
test_with_class_target_file = new FileVMatrix(test_with_class_target_file_name);
test_with_class_target_file->defineSizes(test_with_class_target_file->width(), 0, 0);
cout << test_with_class_target_file_name << " already exist, we are skipping this step." << endl;
}
else
{
compute_target_learner = ::PLearn::deepCopy(compute_target_learner_template);
compute_target_learner->unknown_sales = 0;
compute_target_learner->output_path = output_path;
compute_target_learner->setTrainingSet(test_set, true);
test_with_class_target_file = compute_target_learner->getOutputFile();
}
cout << endl << "****** STEP 2 ******" << endl;
cout << "Shuffling is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 3 ******" << endl;
cout << "For strictly binary variables, various situations arise: zero or non-zero, missing or not-missing, a given value or not, etc..." << endl;
cout << "This step uses FixDond2BinaryVariables to create step3_test_with_binary_fixed.pmat with 0-1 binary variables." << endl;
output_path = expdir+"step3_test_with_binary_fixed";
test_with_binary_fixed_file_name = output_path + ".pmat";
if (isfile(test_with_binary_fixed_file_name))
{
test_with_binary_fixed_file = new FileVMatrix(test_with_binary_fixed_file_name);
test_with_binary_fixed_file->defineSizes(test_with_binary_fixed_file->width(), 0, 0);
cout << test_with_binary_fixed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
fix_binary_variables_learner = ::PLearn::deepCopy(fix_binary_variables_template);
fix_binary_variables_learner->output_path = output_path;
fix_binary_variables_learner->setTrainingSet(test_with_class_target_file, true);
test_with_binary_fixed_file = fix_binary_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 4 ******" << endl;
cout << "This step adds missing indicators variables to each variable with missing values." << endl;
cout << "It uses MissingIndicatorVMatrix to transform step3_test_with_binary_fixed.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 6" << endl;
output_path = expdir+"step6_test_with_dichotomies";
test_with_dichotomies_file_name = output_path + ".pmat";
if (isfile(test_with_dichotomies_file_name))
{
cout << test_with_dichotomies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_with_ind_vmatrix = new MissingIndicatorVMatrix();
test_with_ind_vmatrix->source = test_with_binary_fixed_file;
test_with_ind_vmatrix->train_set = train_with_binary_fixed_file;
test_with_ind_vmatrix->number_of_train_samples_to_use = 30000.0;
test_with_ind_vmatrix->build();
test_with_ind_vmat = test_with_ind_vmatrix;
}
cout << endl << "****** STEP 5 ******" << endl;
cout << "Computing mean, median and mode is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 6 ******" << endl;
cout << "This steps generates as many dichotomized variables as there are significant code values." << endl;
cout << "It uses DichotomizeDond2DiscreteVariables to transform the vmat from step 4 into step6_test_with_dichotomies.pmat" << endl;
if (isfile(test_with_dichotomies_file_name))
{
test_with_dichotomies_file = new FileVMatrix(test_with_dichotomies_file_name);
test_with_dichotomies_file->defineSizes(test_with_dichotomies_file->width(), 0, 0);
cout << test_with_dichotomies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
dichotomize_discrete_variables_learner = new DichotomizeDond2DiscreteVariables();
dichotomize_discrete_variables_learner->discrete_variable_instructions = discrete_variable_instructions;
dichotomize_discrete_variables_learner->output_path = output_path;
dichotomize_discrete_variables_learner->setTrainingSet(test_with_ind_vmat, true);
test_with_dichotomies_file = dichotomize_discrete_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 7 ******" << endl;
cout << "Dichotomizing mean median and mode is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 8 ******" << endl;
cout << "This step select the desired columns from the test set to create the input records." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_test_with_dichotomies.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 10" << endl;
output_path = expdir+"final_test_input_preprocessed";
test_input_preprocessed_file_name = output_path + ".pmat";
if (isfile(test_input_preprocessed_file_name))
{
cout << test_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
test_with_selected_columns_vmatrix->source = test_with_dichotomies_file;
test_with_selected_columns_vmatrix->fields_partial_match = 0;
test_with_selected_columns_vmatrix->extend_with_missing = 0;
test_with_selected_columns_vmatrix->fields = selected_variables_for_input;
test_with_selected_columns_vmatrix->build();
test_with_selected_columns_vmatrix->defineSizes(test_with_selected_columns_vmatrix->width(), 0, 0);
test_with_selected_columns_vmat = test_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 9 ******" << endl;
cout << "Selecting variables for the mean, median and mode is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 10 ******" << endl;
cout << "This gaussianizes the input records." << endl;
cout << "It uses GaussianizeVMatrix to transform the vmat from step 8" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 12" << endl;
if (isfile(test_input_preprocessed_file_name))
{
cout << test_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_gaussianized_vmatrix = new GaussianizeVMatrix();
test_gaussianized_vmatrix->source = test_with_selected_columns_vmat;
test_gaussianized_vmatrix->train_source = train_with_selected_columns_vmat;
test_gaussianized_vmatrix->threshold_ratio = 1;
test_gaussianized_vmatrix->gaussianize_input = 1;
test_gaussianized_vmatrix->gaussianize_target = 0;
test_gaussianized_vmatrix->gaussianize_weight = 0;
test_gaussianized_vmatrix->gaussianize_extra = 0;
test_gaussianized_vmatrix->excluded_fields = inputs_excluded_from_gaussianization;
test_gaussianized_vmatrix->build();
test_gaussianized_vmat = test_gaussianized_vmatrix;
}
cout << endl << "****** STEP 11 ******" << endl;
cout << "Gaussianizing the mean, meadian and mode is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 12 ******" << endl;
cout << "Finaly, the preprocessed input vectors are store on disk." << endl;
cout << "The vmat from step 10 is converted to final_test_input_preprocessed.pmat." << endl;
if (isfile(test_input_preprocessed_file_name))
{
cout << test_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_input_preprocessed_file = new FileVMatrix(test_input_preprocessed_file_name, test_gaussianized_vmat->length(), test_gaussianized_vmat->fieldNames());
test_input_preprocessed_file->defineSizes(test_gaussianized_vmat->inputsize(), test_gaussianized_vmat->targetsize(), test_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final test preprocessed input records", test_gaussianized_vmat->length());
test_input_preprocessed_vector.resize(test_gaussianized_vmat->width());
for (int test_gaussianized_row = 0; test_gaussianized_row < test_gaussianized_vmat->length(); test_gaussianized_row++)
{
test_gaussianized_vmat->getRow(test_gaussianized_row, test_input_preprocessed_vector);
test_input_preprocessed_file->putRow(test_gaussianized_row, test_input_preprocessed_vector);
pb->update( test_gaussianized_row );
}
delete pb;
}
cout << endl << "****** STEP 13 ******" << endl;
cout << "Saving the final mean, median and mode is not required for the test set, skipped." << endl;
cout << endl << "****** STEP 14 ******" << endl;
cout << "This step select the desired columns from the testing set to create the target records." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_test_with_dichotomies.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 15" << endl;
output_path = expdir+"final_test_target_preprocessed";
test_target_preprocessed_file_name = output_path + ".pmat";
if (isfile(test_target_preprocessed_file_name))
{
cout << test_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_target_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
test_target_with_selected_columns_vmatrix->source = test_with_dichotomies_file;
test_target_with_selected_columns_vmatrix->fields_partial_match = 0;
test_target_with_selected_columns_vmatrix->extend_with_missing = 0;
test_target_with_selected_columns_vmatrix->fields = selected_variables_for_target;
test_target_with_selected_columns_vmatrix->build();
test_target_with_selected_columns_vmatrix->defineSizes(test_target_with_selected_columns_vmatrix->width(), 0, 0);
test_target_with_selected_columns_vmat = test_target_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 15 ******" << endl;
cout << "This gaussianizes the input records." << endl;
cout << "It uses GaussianizeVMatrix to transform the vmat from step 14" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 16" << endl;
if (isfile(test_target_preprocessed_file_name))
{
cout << test_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_target_gaussianized_vmatrix = new GaussianizeVMatrix();
test_target_gaussianized_vmatrix->source = test_target_with_selected_columns_vmat;
test_target_gaussianized_vmatrix->train_source = train_target_with_selected_columns_vmat;
test_target_gaussianized_vmatrix->threshold_ratio = 1;
test_target_gaussianized_vmatrix->gaussianize_input = 1;
test_target_gaussianized_vmatrix->gaussianize_target = 0;
test_target_gaussianized_vmatrix->gaussianize_weight = 0;
test_target_gaussianized_vmatrix->gaussianize_extra = 0;
test_target_gaussianized_vmatrix->excluded_fields = targets_excluded_from_gaussianization;;
test_target_gaussianized_vmatrix->build();
test_target_gaussianized_vmat = test_target_gaussianized_vmatrix;
}
cout << endl << "****** STEP 16 ******" << endl;
cout << "Finaly, the preprocessed input vectors are store on disk." << endl;
cout << "The vmat from step 15 is converted to final_test_target_preprocessed.pmat." << endl;
if (isfile(test_target_preprocessed_file_name))
{
cout << test_target_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
test_target_preprocessed_file = new FileVMatrix(test_target_preprocessed_file_name, test_target_gaussianized_vmat->length(),
test_target_gaussianized_vmat->fieldNames());
test_target_preprocessed_file->defineSizes(test_target_gaussianized_vmat->inputsize(), test_target_gaussianized_vmat->targetsize(),
test_target_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final test preprocessed target records", test_target_gaussianized_vmat->length());
test_target_preprocessed_vector.resize(test_target_gaussianized_vmat->width());
for (int test_gaussianized_row = 0; test_gaussianized_row < test_target_gaussianized_vmat->length(); test_gaussianized_row++)
{
test_target_gaussianized_vmat->getRow(test_gaussianized_row, test_target_preprocessed_vector);
test_target_preprocessed_file->putRow(test_gaussianized_row, test_target_preprocessed_vector);
pb->update( test_gaussianized_row );
}
delete pb;
}
// defining all the variables for the unknown set
PPath unknown_with_class_target_file_name;
VMat unknown_with_class_target_file;
PPath unknown_with_binary_fixed_file_name;
VMat unknown_with_binary_fixed_file;
PPath unknown_with_ind_file_name;
MissingIndicatorVMatrix* unknown_with_ind_vmatrix;
VMat unknown_with_ind_vmat;
PPath unknown_with_dichotomies_file_name;
VMat unknown_with_dichotomies_file;
SelectColumnsVMatrix* unknown_with_selected_columns_vmatrix;
VMat unknown_with_selected_columns_vmat;
GaussianizeVMatrix* unknown_gaussianized_vmatrix;
VMat unknown_gaussianized_vmat;
PPath unknown_input_preprocessed_file_name;
VMat unknown_input_preprocessed_file;
Vec unknown_input_preprocessed_vector;
// managing the unknown set
cout << endl << "********************" << endl;
cout << "In Preprocessing: finally, we format the unknown set" << endl;
cout << endl << "****** STEP 1 ******" << endl;
cout << "The first step groups variables by type, skips untrustworthy variables, and generate class targets" << endl;
cout << "It uses ComputeDond2Target to transform base_unknown.pmat into step1_unknown_with_class_target.pmat" << endl;
output_path = expdir+"step1_unknown_with_class_target";
unknown_with_class_target_file_name = output_path + ".pmat";
if (isfile(unknown_with_class_target_file_name))
{
unknown_with_class_target_file = new FileVMatrix(unknown_with_class_target_file_name);
unknown_with_class_target_file->defineSizes(unknown_with_class_target_file->width(), 0, 0);
cout << unknown_with_class_target_file_name << " already exist, we are skipping this step." << endl;
}
else
{
compute_target_learner = ::PLearn::deepCopy(compute_target_learner_template);
compute_target_learner->unknown_sales = 1;
compute_target_learner->output_path = output_path;
compute_target_learner->setTrainingSet(unknown_set, true);
unknown_with_class_target_file = compute_target_learner->getOutputFile();
}
cout << endl << "****** STEP 2 ******" << endl;
cout << "Shuffling is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 3 ******" << endl;
cout << "For strictly binary variables, various situations arise: zero or non-zero, missing or not-missing, a given value or not, etc..." << endl;
cout << "This step uses FixDond2BinaryVariables to create step3_unknown_with_binary_fixed.pmat with 0-1 binary variables." << endl;
output_path = expdir+"step3_unknown_with_binary_fixed";
unknown_with_binary_fixed_file_name = output_path + ".pmat";
if (isfile(unknown_with_binary_fixed_file_name))
{
unknown_with_binary_fixed_file = new FileVMatrix(unknown_with_binary_fixed_file_name);
unknown_with_binary_fixed_file->defineSizes(unknown_with_binary_fixed_file->width(), 0, 0);
cout << unknown_with_binary_fixed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
fix_binary_variables_learner = ::PLearn::deepCopy(fix_binary_variables_template);
fix_binary_variables_learner->output_path = output_path;
fix_binary_variables_learner->setTrainingSet(unknown_with_class_target_file, true);
unknown_with_binary_fixed_file = fix_binary_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 4 ******" << endl;
cout << "This step adds missing indicators variables to each variable with missing values." << endl;
cout << "It uses MissingIndicatorVMatrix to transform step3_unknown_with_binary_fixed.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 6" << endl;
output_path = expdir+"step6_unknown_with_dichotomies";
unknown_with_dichotomies_file_name = output_path + ".pmat";
if (isfile(unknown_with_dichotomies_file_name))
{
cout << unknown_with_dichotomies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
unknown_with_ind_vmatrix = new MissingIndicatorVMatrix();
unknown_with_ind_vmatrix->source = unknown_with_binary_fixed_file;
unknown_with_ind_vmatrix->train_set = train_with_binary_fixed_file;
unknown_with_ind_vmatrix->number_of_train_samples_to_use = 30000.0;
unknown_with_ind_vmatrix->build();
unknown_with_ind_vmat = unknown_with_ind_vmatrix;
}
cout << endl << "****** STEP 5 ******" << endl;
cout << "Computing mean, median and mode is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 6 ******" << endl;
cout << "This steps generates as many dichotomized variables as there are significant code values." << endl;
cout << "It uses DichotomizeDond2DiscreteVariables to transform the vmat from step 4 into step6_unknown_with_dichotomies.pmat" << endl;
if (isfile(unknown_with_dichotomies_file_name))
{
unknown_with_dichotomies_file = new FileVMatrix(unknown_with_dichotomies_file_name);
unknown_with_dichotomies_file->defineSizes(unknown_with_dichotomies_file->width(), 0, 0);
cout << unknown_with_dichotomies_file_name << " already exist, we are skipping this step." << endl;
}
else
{
dichotomize_discrete_variables_learner = new DichotomizeDond2DiscreteVariables();
dichotomize_discrete_variables_learner->discrete_variable_instructions = discrete_variable_instructions;
dichotomize_discrete_variables_learner->output_path = output_path;
dichotomize_discrete_variables_learner->setTrainingSet(unknown_with_ind_vmat, true);
unknown_with_dichotomies_file = dichotomize_discrete_variables_learner->getOutputFile();
}
cout << endl << "****** STEP 7 ******" << endl;
cout << "Dichotomizing mean median and mode is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 8 ******" << endl;
cout << "This step select the desired columns from the unknown set to create the input records." << endl;
cout << "It uses SelectColumnsVMatrix to transform step6_unknown_with_dichotomies.pmat" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 10" << endl;
output_path = expdir+"final_unknown_input_preprocessed";
unknown_input_preprocessed_file_name = output_path + ".pmat";
if (isfile(unknown_input_preprocessed_file_name))
{
cout << unknown_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
unknown_with_selected_columns_vmatrix = new SelectColumnsVMatrix();
unknown_with_selected_columns_vmatrix->source = unknown_with_dichotomies_file;
unknown_with_selected_columns_vmatrix->fields_partial_match = 0;
unknown_with_selected_columns_vmatrix->extend_with_missing = 0;
unknown_with_selected_columns_vmatrix->fields = selected_variables_for_input;
unknown_with_selected_columns_vmatrix->build();
unknown_with_selected_columns_vmatrix->defineSizes(unknown_with_selected_columns_vmatrix->width(), 0, 0);
unknown_with_selected_columns_vmat = unknown_with_selected_columns_vmatrix;
}
cout << endl << "****** STEP 9 ******" << endl;
cout << "Selecting variables for the mean, median and mode is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 10 ******" << endl;
cout << "This gaussianizes the input records." << endl;
cout << "It uses GaussianizeVMatrix to transform the vmat from step 8" << endl;
cout << "The resulting vitual view is not stored on disk, it is fed as input to step 12" << endl;
if (isfile(unknown_input_preprocessed_file_name))
{
cout << unknown_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
unknown_gaussianized_vmatrix = new GaussianizeVMatrix();
unknown_gaussianized_vmatrix->source = unknown_with_selected_columns_vmat;
unknown_gaussianized_vmatrix->train_source = train_with_selected_columns_vmat;
unknown_gaussianized_vmatrix->threshold_ratio = 1;
unknown_gaussianized_vmatrix->gaussianize_input = 1;
unknown_gaussianized_vmatrix->gaussianize_target = 0;
unknown_gaussianized_vmatrix->gaussianize_weight = 0;
unknown_gaussianized_vmatrix->gaussianize_extra = 0;
unknown_gaussianized_vmatrix->excluded_fields = inputs_excluded_from_gaussianization;
unknown_gaussianized_vmatrix->build();
unknown_gaussianized_vmat = unknown_gaussianized_vmatrix;
}
cout << endl << "****** STEP 11 ******" << endl;
cout << "Gaussianizing the mean, meadian and mode is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 12 ******" << endl;
cout << "Finaly, the preprocessed input vectors are store on disk." << endl;
cout << "The vmat from step 10 is converted to final_unknown_input_preprocessed.pmat." << endl;
if (isfile(unknown_input_preprocessed_file_name))
{
cout << unknown_input_preprocessed_file_name << " already exist, we are skipping this step." << endl;
}
else
{
unknown_input_preprocessed_file = new FileVMatrix(unknown_input_preprocessed_file_name, unknown_gaussianized_vmat->length(), unknown_gaussianized_vmat->fieldNames());
unknown_input_preprocessed_file->defineSizes(unknown_gaussianized_vmat->inputsize(), unknown_gaussianized_vmat->targetsize(), unknown_gaussianized_vmat->weightsize());
pb = new ProgressBar("Saving the final unknown preprocessed input records", unknown_gaussianized_vmat->length());
unknown_input_preprocessed_vector.resize(unknown_gaussianized_vmat->width());
for (int unknown_gaussianized_row = 0; unknown_gaussianized_row < unknown_gaussianized_vmat->length(); unknown_gaussianized_row++)
{
unknown_gaussianized_vmat->getRow(unknown_gaussianized_row, unknown_input_preprocessed_vector);
unknown_input_preprocessed_file->putRow(unknown_gaussianized_row, unknown_input_preprocessed_vector);
pb->update( unknown_gaussianized_row );
}
delete pb;
}
cout << endl << "****** STEP 13 ******" << endl;
cout << "Saving the final mean, median and mode is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 14 ******" << endl;
cout << "Saving the final target preprocessed records is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 15 ******" << endl;
cout << "Saving the final target preprocessed records is not required for the unknown set, skipped." << endl;
cout << endl << "****** STEP 16 ******" << endl;
cout << "Saving the final target preprocessed records is not required for the unknown set, skipped." << endl;
}
| int PLearn::Preprocessing::outputsize | ( | ) | const [virtual] |
SUBCLASS WRITING: override this so that it returns the size of this learner's output, as a function of its inputsize(), targetsize() and set options.
Implements PLearn::PLearner.
Definition at line 959 of file Preprocessing.cc.
{return 0;}
| void PLearn::Preprocessing::train | ( | ) | [virtual] |
*** SUBCLASS WRITING: ***
The role of the train method is to bring the learner up to stage==nstages, updating the stats with training costs measured on-line in the process.
TYPICAL CODE:
static Vec input; // static so we don't reallocate/deallocate memory each time... static Vec target; // (but be careful that static means shared!) input.resize(inputsize()); // the train_set's inputsize() target.resize(targetsize()); // the train_set's targetsize() real weight; if(!train_stats) // make a default stats collector, in case there's none train_stats = new VecStatsCollector(); if(nstages<stage) // asking to revert to a previous stage! forget(); // reset the learner to stage=0 while(stage<nstages) { // clear statistics of previous epoch train_stats->forget(); //... train for 1 stage, and update train_stats, // using train_set->getSample(input, target, weight); // and train_stats->update(train_costs) ++stage; train_stats->finalize(); // finalize statistics for this epoch }
Implements PLearn::PLearner.
Definition at line 955 of file Preprocessing.cc.
{
}
Reimplemented from PLearn::PLearner.
Definition at line 114 of file Preprocessing.h.
The template of the script to generate the class target.
Definition at line 76 of file Preprocessing.h.
Referenced by declareOptions().
| TVec< pair<string, TVec< pair<real, real> > > > PLearn::Preprocessing::discrete_variable_instructions |
The instructions to fix the binary variables in the form of field_name : instruction.
Supported instructions are 9_is_one, not_0_is_one, not_missing_is_one, not_1000_is_one. Variables with no specification will be kept as_is.
Definition at line 84 of file Preprocessing.h.
Referenced by declareOptions().
The template of the script to fix the binary variables.
Definition at line 78 of file Preprocessing.h.
Referenced by declareOptions().
| TVec< pair<string, string> > PLearn::Preprocessing::imputation_spec |
Pairs of instruction of the form field_name : mean | median | mode.
Definition at line 80 of file Preprocessing.h.
Referenced by declareOptions().
The list of variables excluded from the gaussianization step.
Definition at line 90 of file Preprocessing.h.
Referenced by declareOptions().
The list of variables selected as input vector.
Definition at line 86 of file Preprocessing.h.
Referenced by declareOptions().
The list of variables selected as target vector.
Definition at line 88 of file Preprocessing.h.
Referenced by declareOptions().
The list of variables excluded from the gaussianization step.
Definition at line 92 of file Preprocessing.h.
Referenced by declareOptions().
### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //!
The test data set.
Definition at line 72 of file Preprocessing.h.
Referenced by declareOptions().
The unknown data set.
Definition at line 74 of file Preprocessing.h.
Referenced by declareOptions().
 1.7.4
1.7.4