|
PLearn 0.1
|
|
PLearn 0.1
|
The first sentence should be a BRIEF DESCRIPTION of what the class does. More...
#include <GaussianizeVMatrix.h>
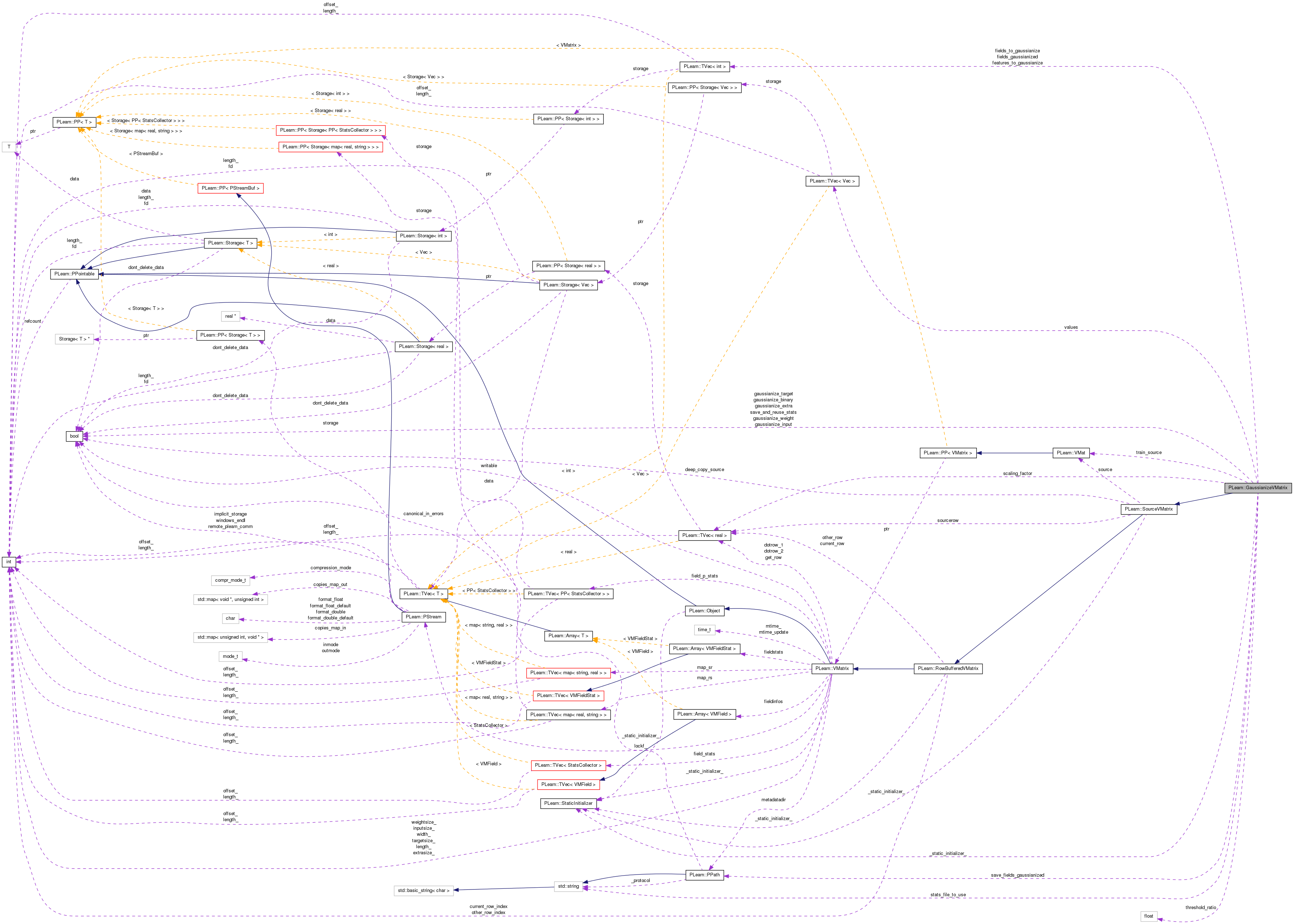
Public Member Functions | |
| GaussianizeVMatrix () | |
| Default constructor. | |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual GaussianizeVMatrix * | deepCopy (CopiesMap &copies) const |
| virtual void | build () |
| Simply calls inherited::build() then build_(). | |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Transforms a shallow copy into a deep copy. | |
| real | unGauss (real value, int col) const |
| return the approximate value of value before being gaussianized. | |
| void | unGauss (Vec &values, Vec &ret, int col) const |
Static Public Member Functions | |
| static string | _classname_ () |
| Declares name and deepCopy methods. | |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| bool | gaussianize_input |
| ### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //! | |
| bool | gaussianize_target |
| bool | gaussianize_weight |
| bool | gaussianize_extra |
| bool | gaussianize_binary |
| real | threshold_ratio |
| bool | save_and_reuse_stats |
| VMat | train_source |
| string | stats_file_to_use |
| TVec< int > | fields_to_gaussianize |
| PPath | save_fields_gaussianized |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Protected Member Functions | |
| virtual void | getNewRow (int i, const Vec &v) const |
| Fill the vector 'v' with the content of the i-th row. | |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares the class options. | |
| static void | declareMethods (RemoteMethodMap &rmm) |
| Declare the methods that are remote-callable. | |
Protected Attributes | |
| TVec< int > | features_to_gaussianize |
| List of features that need to be Gaussianized. | |
| TVec< int > | fields_gaussianized |
| Vec | scaling_factor |
| Scaling factor to map the rank to [0,1]. | |
| TVec< Vec > | values |
| The j-th element is the list of values appearing in the variable features_to_gaussianize[j]. | |
Private Types | |
| typedef SourceVMatrix | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
| virtual void | setMetaDataDir (const PPath &the_metadatadir) |
| Also sets the source's meta-data dir if it's not already set. | |
| real | remote_unGauss (real input, int col) const |
| Version of unGauss(vec,vec,int) that's called by RMI. | |
| Vec | remote_unGauss_vec (Vec inputs, int col) const |
| Version of unGauss(vec,vec,int) that's called by RMI. | |
| void | append_col_to_gaussianize (int col, StatsCollector stat) |
The first sentence should be a BRIEF DESCRIPTION of what the class does.
Place the rest of the class programmer documentation here. Doxygen supports Javadoc-style comments. See http://www.doxygen.org/manual.html
Definition at line 57 of file GaussianizeVMatrix.h.
typedef SourceVMatrix PLearn::GaussianizeVMatrix::inherited [private] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 59 of file GaussianizeVMatrix.h.
| PLearn::GaussianizeVMatrix::GaussianizeVMatrix | ( | ) |
Default constructor.
Definition at line 81 of file GaussianizeVMatrix.cc.
:
gaussianize_input(true),
gaussianize_target(false),
gaussianize_weight(false),
gaussianize_extra(false),
gaussianize_binary(false),
threshold_ratio(10),
save_and_reuse_stats(true)
{}
| string PLearn::GaussianizeVMatrix::_classname_ | ( | ) | [static] |
Declares name and deepCopy methods.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| OptionList & PLearn::GaussianizeVMatrix::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| RemoteMethodMap & PLearn::GaussianizeVMatrix::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| Object * PLearn::GaussianizeVMatrix::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| StaticInitializer GaussianizeVMatrix::_static_initializer_ & PLearn::GaussianizeVMatrix::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| void PLearn::GaussianizeVMatrix::append_col_to_gaussianize | ( | int | col, |
| StatsCollector | stat | ||
| ) | [private] |
Definition at line 230 of file GaussianizeVMatrix.cc.
References PLearn::TVec< T >::append(), features_to_gaussianize, PLearn::StatsCollector::getApproximateCounts(), PLearn::TVec< T >::lastElement(), PLearn::TVec< T >::resize(), and values.
Referenced by setMetaDataDir().
{
values.append(Vec());
Vec& values_j = values.lastElement();
features_to_gaussianize.append(col);
map<real, StatsCollectorCounts>::const_iterator it, it_dummy;
// Note that we obtain the approximate counts, so that almost equal
// values have been merged together already.
map<real,StatsCollectorCounts>* count_map =
stat.getApproximateCounts();
values_j.resize(0,count_map->size());
// We use a dummy iterator to get rid of the last element in the
// map, which is the max real value.
it_dummy = count_map->begin();
it_dummy++;
for (it = count_map->begin(); it_dummy != count_map->end();
it++, it_dummy++)
{
values_j.append(it->first);
}
}


| void PLearn::GaussianizeVMatrix::build | ( | ) | [virtual] |
Simply calls inherited::build() then build_().
Reimplemented from PLearn::SourceVMatrix.
Definition at line 169 of file GaussianizeVMatrix.cc.
References PLearn::SourceVMatrix::build(), and build_().
Referenced by PLearn::Preprocessing::manageTrainTestUnknownSets().
{
inherited::build();
build_();
}
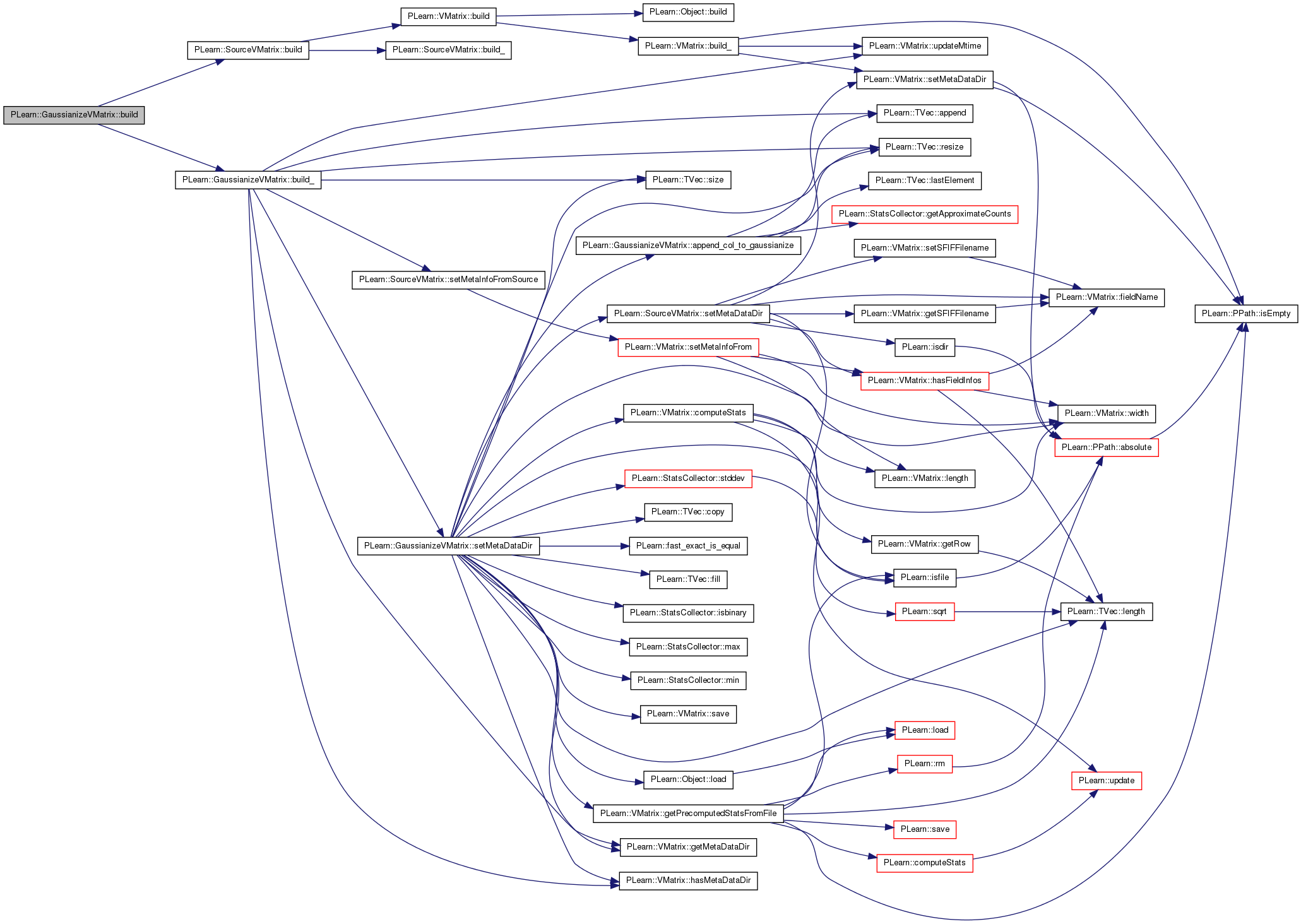

| void PLearn::GaussianizeVMatrix::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 178 of file GaussianizeVMatrix.cc.
References PLearn::TVec< T >::append(), features_to_gaussianize, gaussianize_extra, gaussianize_input, gaussianize_target, gaussianize_weight, PLearn::VMatrix::getMetaDataDir(), PLearn::VMatrix::hasMetaDataDir(), PLCHECK, PLearn::TVec< T >::resize(), save_and_reuse_stats, setMetaDataDir(), PLearn::SourceVMatrix::setMetaInfoFromSource(), PLearn::TVec< T >::size(), PLearn::SourceVMatrix::source, stats_file_to_use, train_source, PLearn::VMatrix::updateMtime(), and values.
Referenced by build().
{
if (!source)
return;
if (train_source) {
source->compatibleSizeError(train_source,
"In GaussianizeVMatrix::build_ -"
" The source and the train_source"
" option are not compatible.");
}
VMat the_source = train_source ? train_source : source;
PLCHECK( the_source->inputsize() >= 0 && the_source->targetsize() >= 0 &&
the_source->weightsize() >= 0 && the_source->extrasize() >= 0 );
// We set the mtime to remove the warning of Mtime=0
if(train_source)
updateMtime(train_source);
updateMtime(source);
// Find which dimensions to Gaussianize.
features_to_gaussianize.resize(0);
int col = 0;
if (gaussianize_input)
features_to_gaussianize.append(
TVec<int>(col, col + the_source->inputsize() - 1, 1));
col += the_source->inputsize();
if (gaussianize_target)
features_to_gaussianize.append(
TVec<int>(col, col + the_source->targetsize() - 1, 1));
col += the_source->targetsize();
if (gaussianize_weight)
features_to_gaussianize.append(
TVec<int>(col, col + the_source->weightsize() - 1, 1));
col += the_source->weightsize();
if (gaussianize_extra)
features_to_gaussianize.append(
TVec<int>(col, col + the_source->extrasize() - 1, 1));
col += the_source->extrasize();
// Obtain meta information from source.
setMetaInfoFromSource();
if((the_source->hasMetaDataDir()||hasMetaDataDir()||!stats_file_to_use.empty()||!save_and_reuse_stats) && values.size()==0)
setMetaDataDir(getMetaDataDir());
}
| string PLearn::GaussianizeVMatrix::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| void PLearn::GaussianizeVMatrix::declareMethods | ( | RemoteMethodMap & | rmm | ) | [static, protected] |
Declare the methods that are remote-callable.
Reimplemented from PLearn::VMatrix.
Definition at line 471 of file GaussianizeVMatrix.cc.
References PLearn::SourceVMatrix::_getRemoteMethodMap_(), PLearn::declareMethod(), PLearn::RemoteMethodMap::inherited(), remote_unGauss(), and remote_unGauss_vec().
{
// Insert a backpointer to remote methods; note that this is different from
// declareOptions().
rmm.inherited(inherited::_getRemoteMethodMap_());
declareMethod(
rmm, "unGauss", &GaussianizeVMatrix::remote_unGauss,
(BodyDoc("Revert the gaussinisation done."),
ArgDoc ("value", "The value to revert."),
ArgDoc ("j", "The column of the value.")));
declareMethod(
rmm, "unGauss2", &GaussianizeVMatrix::remote_unGauss_vec,
(BodyDoc("Revert the gaussinisation done."),
ArgDoc ("values", "A vector of values to revert."),
ArgDoc ("j", "The column of the value.")));
}
| void PLearn::GaussianizeVMatrix::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares the class options.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 94 of file GaussianizeVMatrix.cc.
References PLearn::OptionBase::buildoption, PLearn::declareOption(), PLearn::SourceVMatrix::declareOptions(), features_to_gaussianize, fields_to_gaussianize, gaussianize_binary, gaussianize_extra, gaussianize_input, gaussianize_target, gaussianize_weight, PLearn::OptionBase::learntoption, PLearn::OptionBase::nosave, save_and_reuse_stats, save_fields_gaussianized, stats_file_to_use, threshold_ratio, train_source, and values.
{
declareOption(ol, "threshold_ratio", &GaussianizeVMatrix::threshold_ratio,
OptionBase::buildoption,
"A source's feature will be Gaussianized when the following holds:\n"
"(max - min) / stddev > threshold_ratio.");
declareOption(ol, "gaussianize_input",
&GaussianizeVMatrix::gaussianize_input,
OptionBase::buildoption,
"Whether or not to Gaussianize the input part.");
declareOption(ol, "gaussianize_target",
&GaussianizeVMatrix::gaussianize_target,
OptionBase::buildoption,
"Whether or not to Gaussianize the target part.");
declareOption(ol, "gaussianize_weight",
&GaussianizeVMatrix::gaussianize_weight,
OptionBase::buildoption,
"Whether or not to Gaussianize the weight part.");
declareOption(ol, "gaussianize_extra",
&GaussianizeVMatrix::gaussianize_extra,
OptionBase::buildoption,
"Whether or not to Gaussianize the extra part.");
declareOption(ol, "save_and_reuse_stats",
&GaussianizeVMatrix::save_and_reuse_stats,
OptionBase::buildoption,
"If true, will save and reuse the stats of the source.");
declareOption(ol, "gaussianize_binary",
&GaussianizeVMatrix::gaussianize_binary,
OptionBase::buildoption,
"Whether or not to Gaussianize binary variable.");
declareOption(ol, "train_source", &GaussianizeVMatrix::train_source,
OptionBase::buildoption,
"An optional VMat that will be used instead of 'source' to compute\n"
"the transformation parameters from the distribution statistics.");
declareOption(ol, "fields_to_gaussianize",
&GaussianizeVMatrix::fields_to_gaussianize,
OptionBase::buildoption,
"The fields that we want to be gaussianized.");
declareOption(ol, "stats_file_to_use",
&GaussianizeVMatrix::stats_file_to_use,
OptionBase::buildoption,
"The filename of the statistics to use instead of the"
" train_source.");
declareOption(ol, "save_fields_gaussianized",
&GaussianizeVMatrix::save_fields_gaussianized,
OptionBase::buildoption,
"A path where we will save the fields selected to be gaussianized.");
declareOption(ol, "features_to_gaussianize",
&GaussianizeVMatrix::features_to_gaussianize,
OptionBase::learntoption,
"The columsn that will be gaussianized.");
declareOption(ol, "values",
&GaussianizeVMatrix::values,
OptionBase::learntoption|OptionBase::nosave,
"The values used to gaussinaze.");
// Now call the parent class' declareOptions
inherited::declareOptions(ol);
}
| static const PPath& PLearn::GaussianizeVMatrix::declaringFile | ( | ) | [inline, static] |
| GaussianizeVMatrix * PLearn::GaussianizeVMatrix::deepCopy | ( | CopiesMap & | copies | ) | const [virtual] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
Fill the vector 'v' with the content of the i-th row.
v is assumed to be the right size. ### This function must be overridden in your class
Reimplemented from PLearn::SourceVMatrix.
Definition at line 351 of file GaussianizeVMatrix.cc.
References PLearn::fast_gauss_01_quantile(), features_to_gaussianize, PLearn::is_missing(), j, PLearn::TVec< T >::lastElement(), PLearn::TVec< T >::length(), PLearn::max(), PLearn::min(), n, PLASSERT, PLERROR, PLearn::TVec< T >::size(), PLearn::SourceVMatrix::source, and values.
{
if(values.size()==0 && features_to_gaussianize.size()>0)
PLERROR("In GaussianizeVMatrix::getNewRow() - We don't have been build correctly. Try to set a metadatadir or set save_and_reuse_stats=0.");
PLASSERT( source );
source->getRow(i, v);
for (int k = 0; k < features_to_gaussianize.length(); k++) {
int j = features_to_gaussianize[k];
real current_val = v[j];
if (is_missing(current_val))
continue;
// Find closest values in the training data.
Vec& values_j = values[k];
real interpol;
if (current_val < values_j[0]) {
// Smaller than the minimum.
interpol = 0;
} else if (current_val > values_j.lastElement()) {
// Higher than the maximum.
interpol = 1;
} else {
int min = 0;
int max = values_j.length() - 1;
while (max - min > 1) {
int mid = (max + min) / 2;
real mid_val = values_j[mid];
if (current_val < mid_val)
max = mid;
else if (current_val > mid_val)
min = mid;
else {
// Found the exact value.
min = max = mid;
}
}
if (min == max)
interpol = min;
else {
PLASSERT( max - min == 1 );
interpol = (current_val - values_j[min]) /
(values_j[max] - values_j[min]) + min;
PLASSERT( !is_missing(interpol) );
}
}
interpol /= (values_j.length() - 1);
PLASSERT( interpol >= 0 && interpol <= 1 );
// The expectation of the minimum and maximum of n numbers taken from a
// uniform(0,1) distribution are respectively 1/n+1 and n/n+1: we shift
// and rescale 'interpol' to be in [1/n+1, n/n+1] before using the
// inverse of the Gaussian cumulative function.
real n = values_j.length();
interpol = (n - 1) / (n + 1) * interpol + 1 / (n + 1);
v[j] = fast_gauss_01_quantile(interpol);
}
}
| OptionList & PLearn::GaussianizeVMatrix::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| OptionMap & PLearn::GaussianizeVMatrix::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| RemoteMethodMap & PLearn::GaussianizeVMatrix::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::SourceVMatrix.
Definition at line 76 of file GaussianizeVMatrix.cc.
| void PLearn::GaussianizeVMatrix::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Transforms a shallow copy into a deep copy.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 410 of file GaussianizeVMatrix.cc.
References PLearn::deepCopyField(), PLearn::SourceVMatrix::makeDeepCopyFromShallowCopy(), and train_source.
{
inherited::makeDeepCopyFromShallowCopy(copies);
deepCopyField(train_source, copies);
//features_to_gaussianize?
//scaling_factor?
//values?
}

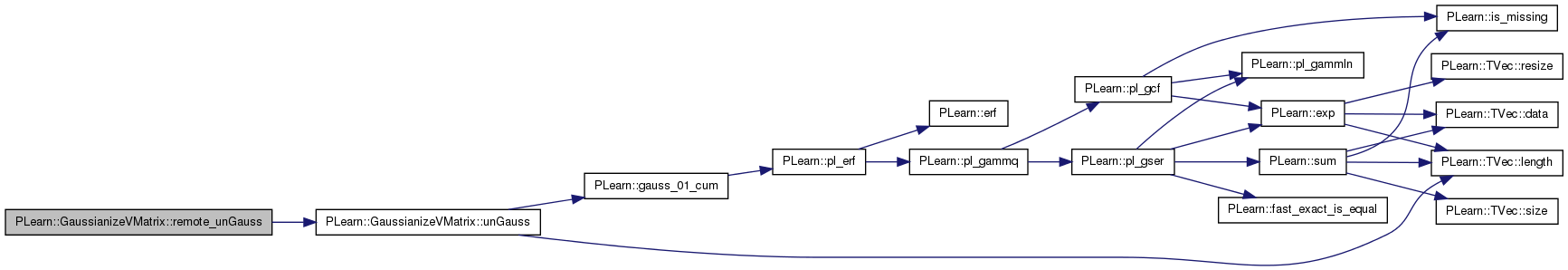
Version of unGauss(vec,vec,int) that's called by RMI.
Definition at line 455 of file GaussianizeVMatrix.cc.
References unGauss().
Referenced by declareMethods().
{
return unGauss(value,col);
}

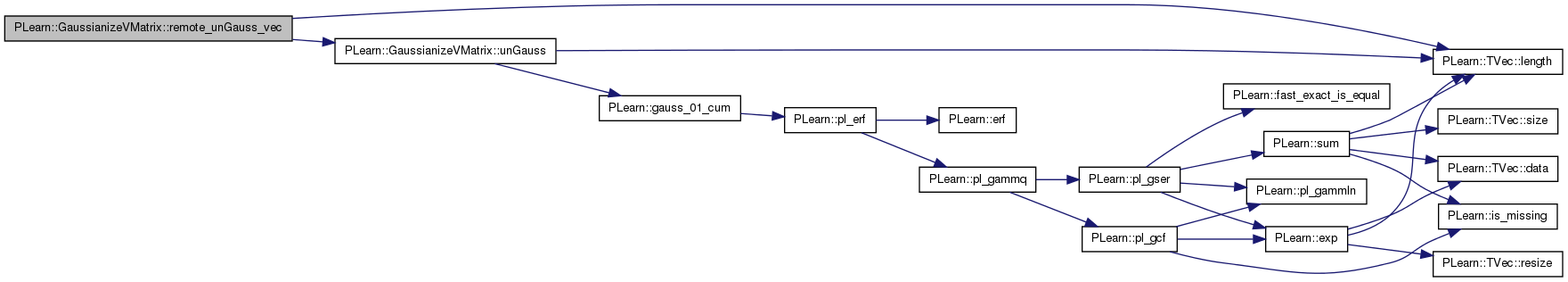
Version of unGauss(vec,vec,int) that's called by RMI.
Definition at line 461 of file GaussianizeVMatrix.cc.
References PLearn::TVec< T >::length(), and unGauss().
Referenced by declareMethods().

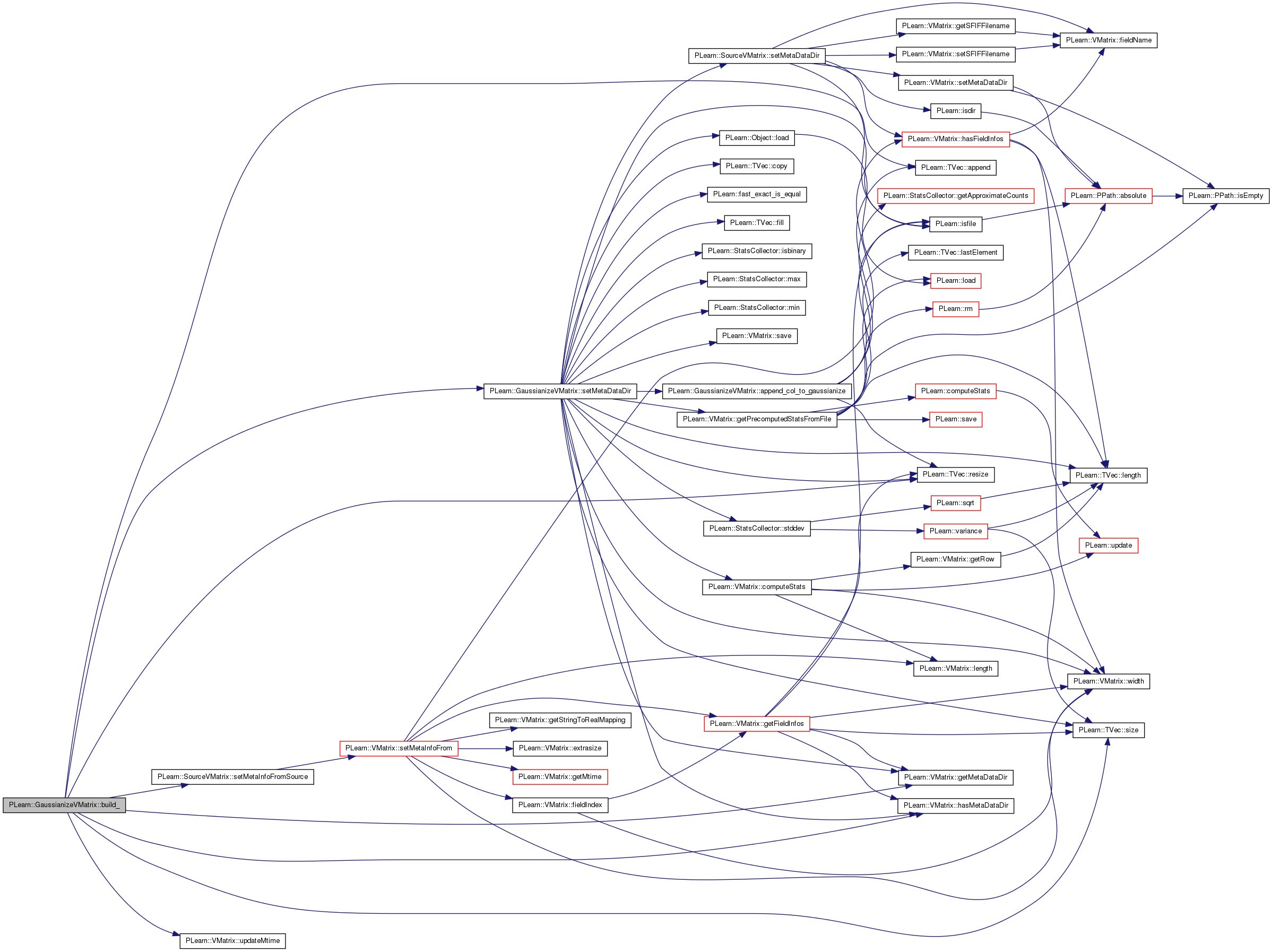
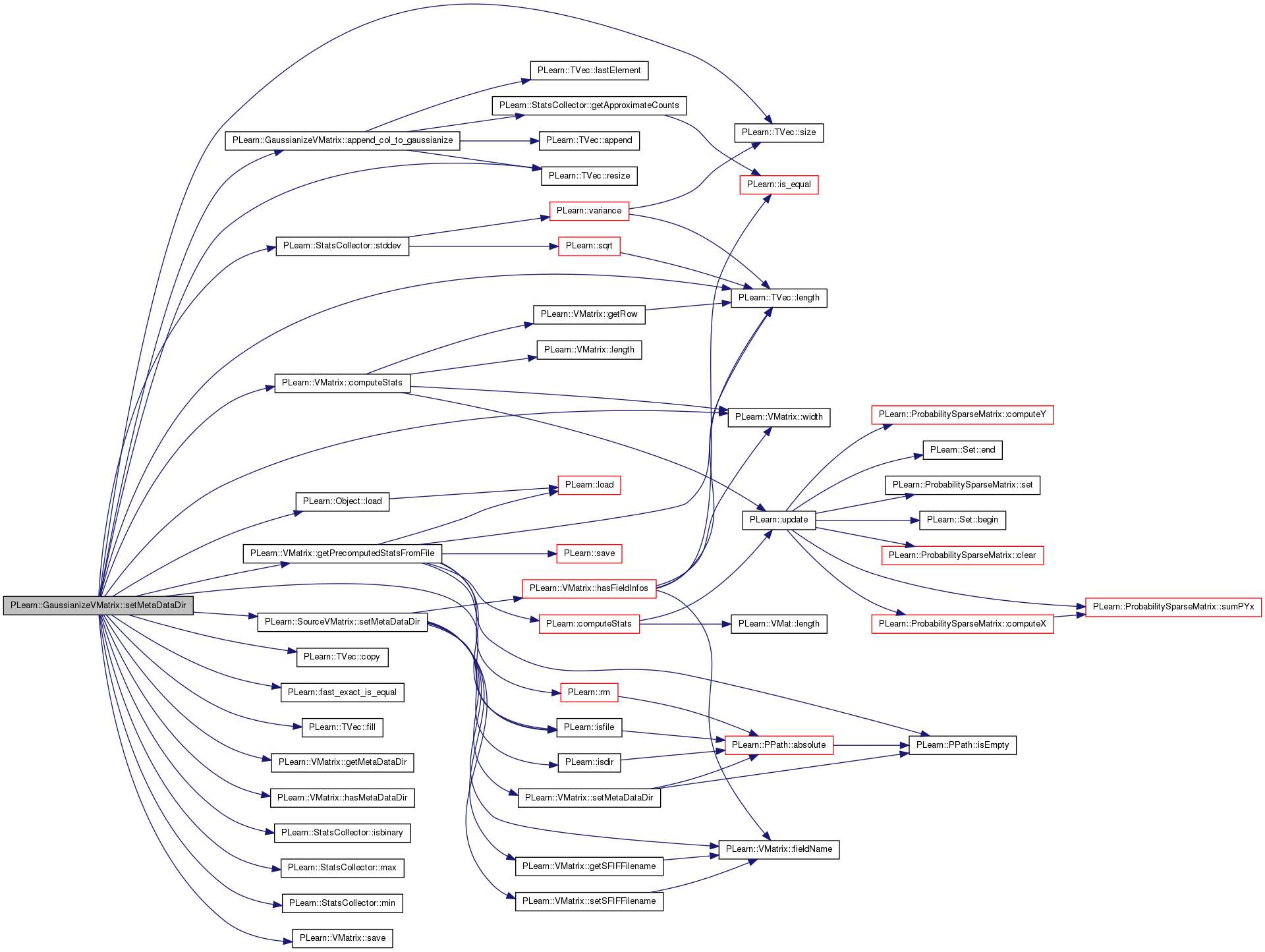
| void PLearn::GaussianizeVMatrix::setMetaDataDir | ( | const PPath & | the_metadatadir | ) | [private, virtual] |
Also sets the source's meta-data dir if it's not already set.
If there are fields that have no corresponding .smap .notes or .binning info files but the source has those files for a field with the same name, then those of the source will be set also for this vmatrix.
Reimplemented from PLearn::SourceVMatrix.
Definition at line 254 of file GaussianizeVMatrix.cc.
References append_col_to_gaussianize(), PLearn::VMatrix::computeStats(), PLearn::TVec< T >::copy(), PLearn::fast_exact_is_equal(), features_to_gaussianize, fields_gaussianized, fields_to_gaussianize, PLearn::TVec< T >::fill(), gaussianize_binary, PLearn::VMatrix::getMetaDataDir(), PLearn::VMatrix::getPrecomputedStatsFromFile(), PLearn::VMatrix::hasMetaDataDir(), i, PLearn::StatsCollector::isbinary(), PLearn::isfile(), j, PLearn::TVec< T >::length(), PLearn::Object::load(), PLearn::StatsCollector::max(), PLearn::StatsCollector::min(), PLERROR, PLWARNING, PLearn::TVec< T >::resize(), PLearn::VMatrix::save(), save_and_reuse_stats, save_fields_gaussianized, PLearn::SourceVMatrix::setMetaDataDir(), PLearn::TVec< T >::size(), PLearn::SourceVMatrix::source, stats_file_to_use, PLearn::StatsCollector::stddev(), threshold_ratio, train_source, values, and PLearn::VMatrix::width().
Referenced by build_().
{
if(!the_metadatadir.empty())
inherited::setMetaDataDir(the_metadatadir);
if(features_to_gaussianize.size()==0)
return;
VMat the_source = train_source ? train_source : source;
if((!the_source->hasMetaDataDir() && stats_file_to_use.empty()) && save_and_reuse_stats)
PLERROR("In GaussianizeVMatrix::setMetaDataDir() - the "
" train_source, source or this VMatrix should have a metadata directory or save_and_reuse_stats must be false");
//to save the stats their must be a metadatadir
if(!the_source->hasMetaDataDir() && hasMetaDataDir()){
if (train_source)
the_source->setMetaDataDir(getMetaDataDir()+"train_source");
else
the_source->setMetaDataDir(getMetaDataDir()+"source");
}
TVec<StatsCollector> stats;
if(!stats_file_to_use.empty()){
if(!isfile(stats_file_to_use))
PLERROR("In GaussianizeVMatrix::setMetaDataDir() - "
"stats_file_to_use = '%s' is not a file.",
stats_file_to_use.c_str());
PLearn::load(stats_file_to_use, stats);
} else if(save_and_reuse_stats)
stats = the_source->
getPrecomputedStatsFromFile("stats_gaussianizeVMatrix.psave", -1, true);
else
stats = PLearn::computeStats(the_source, -1, true);
if(fields_to_gaussianize.size()>0){
if(fields_to_gaussianize.size()>width())
PLERROR("In GaussianizeVMatrix::setMetaDataDir() - "
"More fields in fields_to_gaussianize then the weidth()");
for(int i=0;i<fields_to_gaussianize.size();i++){
int field=fields_to_gaussianize[i];
if(field>=width() || field<0)
PLERROR("In GaussianizeVMatrix::setMetaDataDir() - "
"bad fields number (%d) in fields_to_gaussianize!",
field);
}
features_to_gaussianize.resize(0,fields_to_gaussianize.length());
values.resize(0);
int last_j=-1;
for (int i = 0; i < fields_to_gaussianize.length(); i++) {
int j = fields_to_gaussianize[i];
StatsCollector& stat = stats[j];
if(last_j+1!=j)
for(int k=last_j+1;k<j;k++){
//to keep the total memory used lower faster.
stats[k].forget();
}
append_col_to_gaussianize(j,stat);
stats[j].forget();//to keep the total memory used lower.
}
}else{
// See which dimensions violate the Gaussian assumption and will be
// actually Gaussianized, and store the corresponding list of values.
TVec<int> candidates = features_to_gaussianize.copy();
features_to_gaussianize.resize(0);
values.resize(0);
for (int i = 0; i < candidates.length(); i++) {
int j = candidates[i];
StatsCollector& stat = stats[j];
if (fast_exact_is_equal(stat.stddev(), 0)){
//we don't gaussianize
}else if (!gaussianize_binary && stat.isbinary()) {
//we don't gaussianize
}else if ((stat.max() - stat.min()) > threshold_ratio * stat.stddev()) {
append_col_to_gaussianize(j,stat);
}
stats[j].forget();//to keep the total memory used lower.
}
}
fields_gaussianized.resize(width());
fields_gaussianized.fill(-1);
for(int i=0;i<features_to_gaussianize.size();i++)
fields_gaussianized[features_to_gaussianize[i]]=i;
if(!save_fields_gaussianized.empty()){
PLearn::save(save_fields_gaussianized,features_to_gaussianize);
}
if(features_to_gaussianize.size()==0)
PLWARNING("GaussianizeVMatrix::build_() 0 variable was gaussianized");
}

Definition at line 438 of file GaussianizeVMatrix.cc.
References fields_gaussianized, PLearn::gauss_01_cum(), i, j, PLearn::TVec< T >::length(), and values.
{
int k=fields_gaussianized[j];
if(k<0)
ret<<inputs;//was not gaussianized
for(int i=0;i<inputs.size();i++){
real value = inputs[i];
real interpol = gauss_01_cum(value);
Vec& values_j = values[k];
int idx=int(interpol*values_j.length());
ret[i]=values_j[idx];
}
}
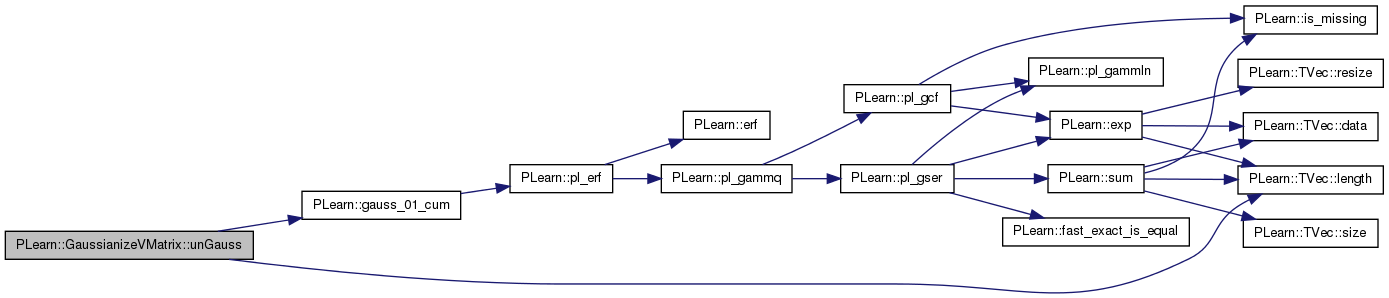
return the approximate value of value before being gaussianized.
Definition at line 423 of file GaussianizeVMatrix.cc.
References fields_gaussianized, PLearn::gauss_01_cum(), j, PLearn::TVec< T >::length(), and values.
Referenced by remote_unGauss(), and remote_unGauss_vec().
{
int k=fields_gaussianized[j];
if(k<0)
return input;//was not gaussianized
real interpol = gauss_01_cum(input);
Vec& values_j = values[k];
int idx=int(interpol*values_j.length());
return values_j[idx];
}

Reimplemented from PLearn::SourceVMatrix.
Definition at line 93 of file GaussianizeVMatrix.h.
TVec<int> PLearn::GaussianizeVMatrix::features_to_gaussianize [protected] |
List of features that need to be Gaussianized.
Definition at line 109 of file GaussianizeVMatrix.h.
Referenced by append_col_to_gaussianize(), build_(), declareOptions(), getNewRow(), and setMetaDataDir().
TVec<int> PLearn::GaussianizeVMatrix::fields_gaussianized [protected] |
Definition at line 111 of file GaussianizeVMatrix.h.
Referenced by setMetaDataDir(), and unGauss().
Definition at line 76 of file GaussianizeVMatrix.h.
Referenced by declareOptions(), and setMetaDataDir().
Definition at line 71 of file GaussianizeVMatrix.h.
Referenced by declareOptions(), and setMetaDataDir().
Definition at line 70 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and PLearn::Preprocessing::manageTrainTestUnknownSets().
### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //!
Definition at line 67 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and PLearn::Preprocessing::manageTrainTestUnknownSets().
Definition at line 68 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and PLearn::Preprocessing::manageTrainTestUnknownSets().
Definition at line 69 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and PLearn::Preprocessing::manageTrainTestUnknownSets().
Definition at line 73 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and setMetaDataDir().
Definition at line 77 of file GaussianizeVMatrix.h.
Referenced by declareOptions(), and setMetaDataDir().
Vec PLearn::GaussianizeVMatrix::scaling_factor [protected] |
Scaling factor to map the rank to [0,1].
Definition at line 114 of file GaussianizeVMatrix.h.
Definition at line 75 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), and setMetaDataDir().
Definition at line 72 of file GaussianizeVMatrix.h.
Referenced by declareOptions(), PLearn::Preprocessing::manageTrainTestUnknownSets(), and setMetaDataDir().
Definition at line 74 of file GaussianizeVMatrix.h.
Referenced by build_(), declareOptions(), makeDeepCopyFromShallowCopy(), PLearn::Preprocessing::manageTrainTestUnknownSets(), and setMetaDataDir().
TVec<Vec> PLearn::GaussianizeVMatrix::values [protected] |
The j-th element is the list of values appearing in the variable features_to_gaussianize[j].
Definition at line 118 of file GaussianizeVMatrix.h.
Referenced by append_col_to_gaussianize(), build_(), declareOptions(), getNewRow(), setMetaDataDir(), and unGauss().
 1.7.4
1.7.4