|
PLearn 0.1
|
|
PLearn 0.1
|
#include <ProbSparseMatrix.h>
Public Member Functions | |
| ProbSparseMatrix (int n_rows=0, int n_cols=0, string name="pXY", int mode=ROW_WISE, bool double_access=false) | |
| void | incr (int i, int j, real inc=1.0, bool warning=true) |
| void | set (int i, int j, real value, bool warning=true) |
| bool | checkCondProbIntegrity () |
| bool | checkJointProbIntegrity () |
| void | normalizeCond (ProbSparseMatrix &nXY, bool clear_nXY=false) |
| void | normalizeJoint (ProbSparseMatrix &nXY, bool clear_nXY=false) |
Normalize the matrix nXY as a joint probability matrix 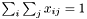 . . | |
| void | normalizeCond () |
| void | normalizeJoint () |
| string | getClassName () const |
| void | iterativeProportionalFittingStep (ProbSparseMatrix &p, Vec &lineMarginal, Vec &colMarginal) |
| real | euclidianDistance (ProbSparseMatrix &p) |
| euclidian distance between two sparse matrices (of same dimensions) | |
| void | add (ProbSparseMatrix &p, ProbSparseMatrix &q) |
Definition at line 45 of file ProbSparseMatrix.h.
| PLearn::ProbSparseMatrix::ProbSparseMatrix | ( | int | n_rows = 0, |
| int | n_cols = 0, |
||
| string | name = "pXY", |
||
| int | mode = ROW_WISE, |
||
| bool | double_access = false |
||
| ) |
Definition at line 41 of file ProbSparseMatrix.cc.
: DoubleAccessSparseMatrix<real>(n_rows, n_cols, name, mode, double_access) { }
| void PLearn::ProbSparseMatrix::add | ( | ProbSparseMatrix & | p, |
| ProbSparseMatrix & | q | ||
| ) |
Definition at line 374 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< T >::exists(), PLearn::DoubleAccessSparseMatrix< T >::get(), PLearn::DoubleAccessSparseMatrix< T >::getCol(), PLearn::DoubleAccessSparseMatrix< T >::getHeight(), PLearn::DoubleAccessSparseMatrix< T >::getRow(), PLearn::DoubleAccessSparseMatrix< T >::getWidth(), i, j, PLearn::DoubleAccessSparseMatrix< real >::mode, PLERROR, and ROW_WISE.
{
real val;
if(p.getHeight()!=q.getHeight() || p.getWidth() != q.getWidth()) PLERROR("euclidianDistance: matrix dimensions do not match ");
if (mode == ROW_WISE){
// go thru the first matrix
for (int i = 0; i < p.getHeight(); i++){
map<int, real>& row_i = p.getRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
val = it->second+q.get(i,it->first);
set(i,it->first,val);
}
}
// go thru the second one
for (int i = 0; i < q.getHeight(); i++){
map<int, real>& row_i = q.getRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
// if the value exists in the first matrix, it has already been seen
if(!p.exists(i,it->first)){
// no value in the first matrix
set(i,it->first,it->second);
}
}
}
}else{
// go thru the first matrix
for (int j = 0; j < p.getWidth(); j++){
map<int, real>& col_j = p.getCol(j);
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
val = it->second+q.get(it->first,j);
set(it->first,j,val);
}
}
// go thru the second one
for (int j = 0; j < q.getWidth(); j++){
map<int, real>& col_j = q.getCol(j);
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
// if the value exists in the first matrix, it has already been seen
if(!p.exists(it->first,j)){
// no value in the first matrix
set(it->first,j,it->second);
}
}
}
}
}
| bool PLearn::ProbSparseMatrix::checkCondProbIntegrity | ( | ) |
Reimplemented in PLearn::SmoothedProbSparseMatrix, and PLearn::ComplementedProbSparseMatrix.
Definition at line 59 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::cols, PLearn::DoubleAccessSparseMatrix< real >::height, i, if(), j, PLearn::DoubleAccessSparseMatrix< real >::mode, ROW_WISE, PLearn::DoubleAccessSparseMatrix< real >::rows, PLearn::sum(), and PLearn::DoubleAccessSparseMatrix< real >::width.
Referenced by PLearn::GraphicalBiText::check_consitency().
{
real sum = 0.0;
if (mode == ROW_WISE)
{
for (int i = 0; i < height; i++)
{
map<int, real>& row_i = rows[i];
sum = 0.0;
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it)
sum += it->second;
if (fabs(sum - 1.0) > 1e-4 && (sum > 0.0))
return false;
}
return true;
} else
{
for (int j = 0; j < width; j++)
{
map<int, real>& col_j = cols[j];
sum = 0.0;
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it)
sum += it->second;
if (fabs(sum - 1.0) > 1e-4 && (sum > 0.0))
return false;
}
return true;
}
}

| bool PLearn::ProbSparseMatrix::checkJointProbIntegrity | ( | ) |
Definition at line 89 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::sumOfElements().
{
return (fabs(sumOfElements() - 1.0) > 1e-4);
}

| real PLearn::ProbSparseMatrix::euclidianDistance | ( | ProbSparseMatrix & | p | ) |
euclidian distance between two sparse matrices (of same dimensions)
Definition at line 250 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::cols, PLearn::diff(), PLearn::distance(), PLearn::DoubleAccessSparseMatrix< real >::exists(), PLearn::DoubleAccessSparseMatrix< T >::get(), PLearn::DoubleAccessSparseMatrix< T >::getCol(), PLearn::DoubleAccessSparseMatrix< T >::getHeight(), PLearn::DoubleAccessSparseMatrix< T >::getRow(), PLearn::DoubleAccessSparseMatrix< T >::getWidth(), PLearn::DoubleAccessSparseMatrix< real >::height, i, j, PLearn::DoubleAccessSparseMatrix< real >::mode, PLERROR, ROW_WISE, PLearn::DoubleAccessSparseMatrix< real >::rows, PLearn::sqrt(), and PLearn::DoubleAccessSparseMatrix< real >::width.
{
real distance=0;
real diff;
if(p.getHeight()!=height || p.getWidth() != width) PLERROR("euclidianDistance: matrix dimensions do not match ");
if (mode == ROW_WISE){
// go thru the first matrix
for (int i = 0; i < height; i++){
map<int, real>& row_i = rows[i];
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
diff = it->second-p.get(i, it->first);
distance +=sqrt(diff*diff);
}
}
// go thru the second one
for (int i = 0; i < p.getHeight(); i++){
map<int, real>& row_i = p.getRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
// if the value exists in the first matrix, it has already been included in the distance
if(exists(i,it->first))continue;
// no value in the first matrix
diff = p.get(i,it->first);
distance +=sqrt(diff*diff);
}
}
}else{
// go thru the first matrix
for (int j = 0; j < width; j++){
map<int, real>& col_j = cols[j];
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
diff = it->second-p.get(it->first,j);
distance +=sqrt(diff*diff);
}
}
// go thru the second one
for (int j = 0; j < width; j++){
map<int, real>& col_j = p.getCol(j);
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
// if the value exists in the first matrix, it has already been included in the distance
if(exists(it->first,j))continue;
// no value in the first matrix
diff = p.get(it->first,j);
distance +=sqrt(diff*diff);
}
}
}
return(distance);
}
| string PLearn::ProbSparseMatrix::getClassName | ( | ) | const [inline, virtual] |
Reimplemented from PLearn::DoubleAccessSparseMatrix< real >.
Reimplemented in PLearn::SmoothedProbSparseMatrix.
Definition at line 68 of file ProbSparseMatrix.h.
{ return "ProbSparseMatrix"; }
Definition at line 45 of file ProbSparseMatrix.cc.
References PLWARNING.
Referenced by PLearn::GraphicalBiText::compute_BN_likelihood(), PLearn::GraphicalBiText::compute_likelihood(), PLearn::GraphicalBiText::init(), PLearn::GraphicalBiText::init_WSD(), and PLearn::GraphicalBiText::update_WSD_model().
{
if (inc <= 0.0 && warning)
PLWARNING("incrementing value by: %g", inc);
DoubleAccessSparseMatrix<real>::incr(i, j, inc);
}
| void PLearn::ProbSparseMatrix::iterativeProportionalFittingStep | ( | ProbSparseMatrix & | p, |
| Vec & | lineMarginal, | ||
| Vec & | colMarginal | ||
| ) |
Definition at line 301 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::cols, PLearn::endl(), PLearn::DoubleAccessSparseMatrix< T >::getCol(), PLearn::DoubleAccessSparseMatrix< T >::getHeight(), PLearn::DoubleAccessSparseMatrix< T >::getRow(), PLearn::DoubleAccessSparseMatrix< T >::getWidth(), PLearn::DoubleAccessSparseMatrix< real >::height, i, j, PLearn::DoubleAccessSparseMatrix< real >::mode, PLearn::DoubleAccessSparseMatrix< T >::mode, PLERROR, PLWARNING, ROW_WISE, PLearn::DoubleAccessSparseMatrix< real >::rows, PLearn::TVec< T >::size(), PLearn::DoubleAccessSparseMatrix< T >::sumCol(), PLearn::DoubleAccessSparseMatrix< T >::sumRow(), and PLearn::DoubleAccessSparseMatrix< real >::width.
{
real newVal;
real sum_row_i;
real sum_col_j;
if(p.getHeight()!=lineMarginal.size() || p.getWidth() != colMarginal.size()) PLERROR("iterativeProportionalFittingStep: matrix dimension does not match marginal vectors dimensions");
if(p.getHeight()!=height || p.getWidth() != width) PLERROR("iterativeProportionalFittingStep: new matrix dimension does not match old matrix dimensions");
if(p.mode!=mode) PLERROR("iterativeProportionalFittingStep: Matrices access mode must match");
if (mode == ROW_WISE){
Vec sum_col(width);
// First pass
for (int i = 0; i < height; i++){
map<int, real>& row_i = p.getRow(i);
sum_row_i = p.sumRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
if(sum_row_i==0)PLERROR("iterativeProportionalFittingStep: line %d is empty",i);
newVal= it->second*lineMarginal[i]/sum_row_i;
// store sum of column for next step
sum_col[it->first]+=newVal;
set(i,it->first,newVal);
}
}
// Second Pass
for (int i = 0; i < height; i++){
// we use the values set in the matrix at the previous stage
map<int, real>& row_i = rows[i];
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it){
if(sum_col[it->first]==0)PLERROR("iterativeProportionalFittingStep: column %d is empty",i);
newVal= it->second*colMarginal[it->first]/sum_col[it->first];
set(i,it->first,newVal);
}
}
}else{
Vec sum_row(height);
for (int j = 0; j < width; j++){
map<int, real>& col_j = p.getCol(j);
sum_col_j = p.sumCol(j);
if( col_j.begin()!= col_j.end())cout << " " << colMarginal[j]/sum_col_j<< ":";
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
if(sum_col_j==0){
PLWARNING("iterativeProportionalFittingStep: column %d is empty",j);
continue;
}
newVal= it->second*colMarginal[j]/sum_col_j;
cout << " " <<it->second<<"="<<newVal;
sum_row[it->first]+=newVal;
set(it->first,j,newVal);
}
if( col_j.begin()!= col_j.end())cout << endl;
}
cout << endl;
// Second Pass
for (int j = 0; j < width; j++){
// we use the values set in the matrix at the previous stage
map<int, real>& col_j = cols[j];
// cout << " " << lineMarginal[it->first]/sum_row[it->first]<< ":";
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it){
if(sum_row[it->first]==0){
PLWARNING("iterativeProportionalFittingStep: line %d is empty",it->first);
continue;
}
newVal= it->second*lineMarginal[it->first]/sum_row[it->first];
cout << " " <<it->second<<"="<<newVal;
set(it->first,j,newVal);
}
cout << endl;
}
}
}
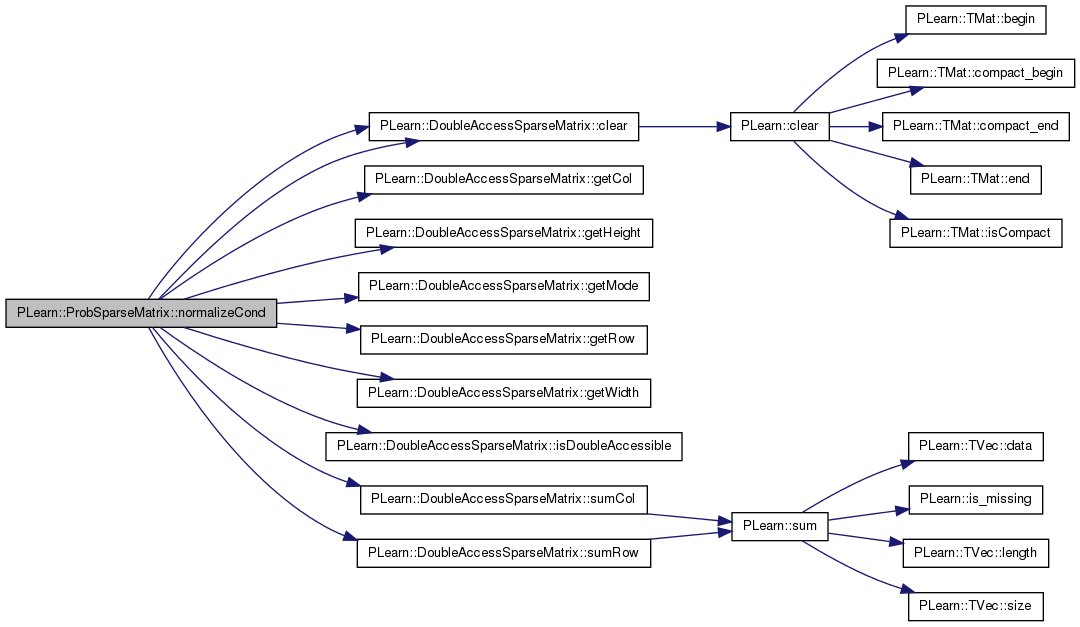
| void PLearn::ProbSparseMatrix::normalizeCond | ( | ProbSparseMatrix & | nXY, |
| bool | clear_nXY = false |
||
| ) |
Definition at line 94 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< T >::clear(), PLearn::DoubleAccessSparseMatrix< real >::clear(), COLUMN_WISE, PLearn::DoubleAccessSparseMatrix< T >::getCol(), PLearn::DoubleAccessSparseMatrix< T >::getHeight(), PLearn::DoubleAccessSparseMatrix< T >::getMode(), PLearn::DoubleAccessSparseMatrix< T >::getRow(), PLearn::DoubleAccessSparseMatrix< T >::getWidth(), i, PLearn::DoubleAccessSparseMatrix< T >::isDoubleAccessible(), j, PLearn::DoubleAccessSparseMatrix< real >::mode, PLERROR, ROW_WISE, PLearn::DoubleAccessSparseMatrix< T >::sumCol(), and PLearn::DoubleAccessSparseMatrix< T >::sumRow().
Referenced by PLearn::GraphicalBiText::compute_likelihood(), and PLearn::GraphicalBiText::init().
{
if (mode == ROW_WISE && (nXY.getMode() == ROW_WISE || nXY.isDoubleAccessible()))
{
clear();
int nXY_height = nXY.getHeight();
for (int i = 0; i < nXY_height; i++)
{
real sum_row_i = nXY.sumRow(i);
map<int, real>& row_i = nXY.getRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it)
{
int j = it->first;
real nij = it->second;
real pij = nij / sum_row_i;
if (pij > 0.0)
set(i, j, pij);
}
}
if (clear_nXY)
nXY.clear();
} else if (mode == COLUMN_WISE && (nXY.getMode() == COLUMN_WISE || nXY.isDoubleAccessible()))
{
clear();
int nXY_width = nXY.getWidth();
for (int j = 0; j < nXY_width; j++)
{
real sum_col_j = nXY.sumCol(j);
map<int, real>& col_j = nXY.getCol(j);
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it)
{
int i = it->first;
real nij = it->second;
real pij = nij / sum_col_j;
if (pij > 0.0)
set(i, j, pij);
}
}
if (clear_nXY)
nXY.clear();
} else
{
PLERROR("pXY and nXY accessibility modes must match");
}
}
| void PLearn::ProbSparseMatrix::normalizeCond | ( | ) |
Definition at line 140 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::cols, PLearn::DoubleAccessSparseMatrix< real >::height, i, j, PLearn::DoubleAccessSparseMatrix< real >::mode, ROW_WISE, PLearn::DoubleAccessSparseMatrix< real >::rows, PLearn::DoubleAccessSparseMatrix< real >::sumCol(), PLearn::DoubleAccessSparseMatrix< real >::sumRow(), and PLearn::DoubleAccessSparseMatrix< real >::width.
{
if (mode == ROW_WISE)
{
for (int i = 0; i < height; i++)
{
real sum_row_i = sumRow(i);
map<int, real>& row_i = rows[i];
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it)
{
int j = it->first;
real nij = it->second;
real pij = nij / sum_row_i;
if (pij > 0.0)
set(i, j, pij);
}
}
} else
{
for (int j = 0; j < width; j++)
{
real sum_col_j = sumCol(j);
map<int, real>& col_j = cols[j];
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it)
{
int i = it->first;
real nij = it->second;
real pij = nij / sum_col_j;
if (pij > 0.0)
set(i, j, pij);
}
}
}
}
| void PLearn::ProbSparseMatrix::normalizeJoint | ( | ProbSparseMatrix & | nXY, |
| bool | clear_nXY = false |
||
| ) |
Normalize the matrix nXY as a joint probability matrix 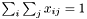 .
.
Definition at line 177 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< T >::clear(), PLearn::DoubleAccessSparseMatrix< real >::clear(), COLUMN_WISE, PLearn::DoubleAccessSparseMatrix< T >::getCol(), PLearn::DoubleAccessSparseMatrix< T >::getHeight(), PLearn::DoubleAccessSparseMatrix< T >::getMode(), PLearn::DoubleAccessSparseMatrix< T >::getRow(), PLearn::DoubleAccessSparseMatrix< T >::getWidth(), i, j, ROW_WISE, and PLearn::DoubleAccessSparseMatrix< T >::sumOfElements().
Referenced by PLearn::GraphicalBiText::init().
{
clear();
real sum_nXY = nXY.sumOfElements();
if (nXY.getMode() == ROW_WISE)
{
int nXY_height = nXY.getHeight();
for (int i = 0; i < nXY_height; i++)
{
map<int, real>& row_i = nXY.getRow(i);
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it)
{
int j = it->first;
real nij = it->second;
real pij = nij / sum_nXY;
if (pij > 0.0)
set(i, j, pij);
}
}
} else if (nXY.getMode() == COLUMN_WISE)
{
int nXY_width = nXY.getWidth();
for (int j = 0; j < nXY_width; j++)
{
map<int, real>& col_j = nXY.getCol(j);
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it)
{
int i = it->first;
real nij = it->second;
real pij = nij / sum_nXY;
if (pij > 0.0)
set(i, j, pij);
}
}
}
if (clear_nXY)
nXY.clear();
}
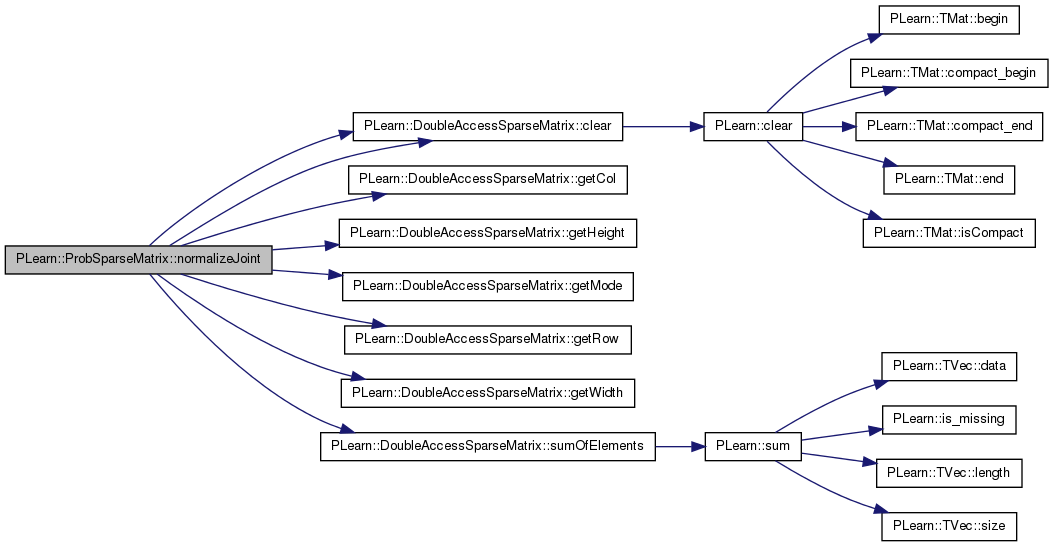

| void PLearn::ProbSparseMatrix::normalizeJoint | ( | ) |
Definition at line 216 of file ProbSparseMatrix.cc.
References PLearn::DoubleAccessSparseMatrix< real >::cols, PLearn::DoubleAccessSparseMatrix< real >::height, i, j, PLearn::DoubleAccessSparseMatrix< real >::mode, ROW_WISE, PLearn::DoubleAccessSparseMatrix< real >::rows, PLearn::DoubleAccessSparseMatrix< real >::sumOfElements(), and PLearn::DoubleAccessSparseMatrix< real >::width.
{
if (mode == ROW_WISE)
{
for (int i = 0; i < height; i++)
{
map<int, real>& row_i = rows[i];
for (map<int, real>::iterator it = row_i.begin(); it != row_i.end(); ++it)
{
int j = it->first;
real nij = it->second;
real pij = nij / sumOfElements();
if (pij > 0.0)
set(i, j, pij);
}
}
} else
{
for (int j = 0; j < width; j++)
{
map<int, real>& col_j = cols[j];
for (map<int, real>::iterator it = col_j.begin(); it != col_j.end(); ++it)
{
int i = it->first;
real nij = it->second;
real pij = nij / sumOfElements();
if (pij > 0.0)
set(i, j, pij);
}
}
}
}

Definition at line 52 of file ProbSparseMatrix.cc.
References PLWARNING.
Referenced by PLearn::GraphicalBiText::compute_likelihood(), PLearn::GraphicalBiText::init(), and PLearn::GraphicalBiText::update_WSD_model().
{
if (value <= 0.0 && warning)
PLWARNING("setting value: %g", value);
DoubleAccessSparseMatrix<real>::set(i, j, value);
}
 1.7.4
1.7.4