| Postdoc |
BAYESIAN INFERENCE AND/OR OPTIMIZATION STRATEGIES FOR
SOME DETECTION, TRACKING AND DECONVOLUTION
PROBLEMS IN MEDICAL IMAGES
Max Mignotte & Jean Meunier
The authors thank INRIA (Institut National de la Recherche en Informatique et Automatique, France) and NSERC (Natural Sciences and Engineering Research Council of Canada) for financial support of this work (postdoctoral grant) respectively from September 1998 until August 1999 and from September 1999 until August 2000. The authors are also grateful to Jean-Paul Soucy and Christian Janicki (CHUM, University of Montreal) for having provided the SPECT images.
| DETECTION AND TRACKING OF ANATOMICAL STRUCTURES USING DEFORMABLE TEMPLATES AND A NOISE MODEL ESTIMATION IN AN ECHOGRAPHIC SEQUENCE |
In this work, we present a new method to shape-based segmentation of deformable anatomical structures in medical images and validate this approach by detecting and tracking the endocardial border in an echographic image sequence. To this end, a global prior knowledge of the endocardial contour is captured by a prototype template with a set of admissible deformations to take into account its inherent natural variability over time. In this approach, the data likelihood model rely on an accurate statistical modeling of the grey level distribution of each class present in the image. The parameters of this distribution mixture are given by a preliminary estimation step which takes into account the distribution shape of each class. Then the tracking problem is stated in a Bayesian framework where it ends up as an optimization problem. This one is then efficiently solved by a genetic algorithm combined with a steepest ascent procedure. This technique has been successfully applied on synthetic images and on a real echocardiographic image sequence. This method seems to be particularly well suited to handle ultrasound images with strong speckle noise on which edge information cannot be exploited. Finally, the local and global minimization procedure we propose is fast, robust and do not require initialization of the template close to the desired solution. Initialization may be defined at random, leading to segmentation and tracking procedure that are completely data driven. (slides)
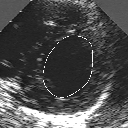
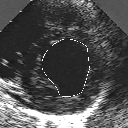
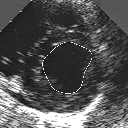
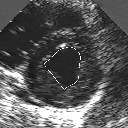
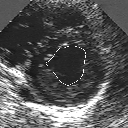
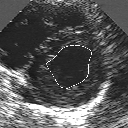
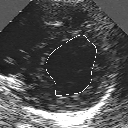
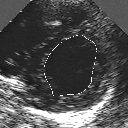
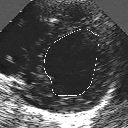
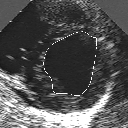
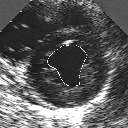
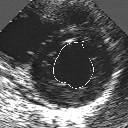
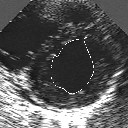
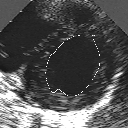
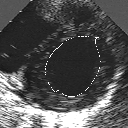
| A MULTISCALE OPTIMIZATION APPROACH FOR THE DYNAMIC CONTOUR-BASED BOUNDARY DETECTION ISSUE |
In this work, we present a new multiscale approach for deformable contour optimization. The method relies on a multigrid minimization method and a coarse-to-fine relaxation algorithm. This approach consists in minimizing a cascade of optimization problems of reduced and increasing complexity instead of considering the minimization problem on the full and original configuration space. Contrary to classical multiresolution algorithms, no reduction of image is applied. The family of defined energy functions are derived from the original (full resolution) objective function, ensuring that the same function is handled at each scale and that the energy decreases at each step of the deformable contour minimization process. The efficiency and the speed of this multiscale optimization strategy is demonstrated in the difficult context of the minimization of a region-based contour energy function ensuring the boundary detection of anatomical structures in ultrasound medical imagery. In this context, the proposed multiscale segmentation method is compared to other classical region-based segmentation approaches such as Maximum Likelihood or Markov Random Field-based segmentation techniques. We also extend this multiscale segmentation strategy to active contour models using a classical edge-based likelihood approach. Finally, time and performance analysis of this approach, compared to the (commonly used) dynamic programming-based optimization procedure, is given and allows to attest the accuracy and the speed of the proposed method. (demo) (slides)
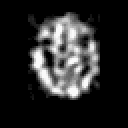
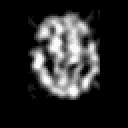
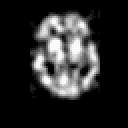
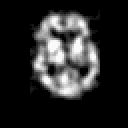
| THREE-DIMENSIONAL BLIND DECONVOLUTION OF SPECT IMAGES |
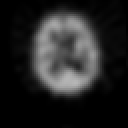
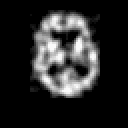
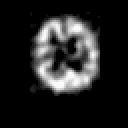
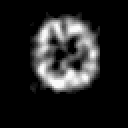
Contrary to other clinical medical imaging techniques, 3D SPECT imagery remains unique in its ability to yield functionally rather than anatomically-based information. This visualization method has become a great help in the diagnostic of cerebrovascular diseases which are the third most common cause of death in the USA and Europe. Nevertheless, 3D SPECT images (or SPECT volume) are very blurred and consequently their interpretation is difficult. In order to improve the spatial resolution of each cross-sectional image from a given SPECT volume and then to facilitate their interpretation by the clinician, we propose to implement and to compare the effectiveness of twelve different existing 2D blind or ``supervised'' deconvolution methods. To this end, we present an accurate distribution mixture parameter estimation procedure which takes into account the diversity of the laws in the distribution mixture of a SPECT image. In our application, parameters of this distribution mixture are efficiently exploited in order to prevent overfitting of the noisy data for the iterative deconvolution techniques without regularization term or to determine the exact support of the object to be restored when this one is needed. Recent 2D blind deconvolution techniques such as the NAS-RIF algorithm combined with this estimation procedure can be efficiently applied in SPECT imagery and yield very promising restoration results.
In order to restore more accurately these 3D SPECT images and to take into account the inter-slice blur in the restoration procedure, we have proposed to extend the abovementioned 2D blind in the deblurring 3D case. This technique requires to find, in a preliminary step, the support of the object to be restored. In this work, we propose to solve this problem thanks to an unsupervised 3D Markovian segmentation technique. This method has been successfully tested on numerous real brain SPECT volumes, yielding very promising restoration results. We have also shown that this 3D blind deconvolution technique gives superior performance than its 2D version. Finally, This 3D blind deconvolution technique combined with this unsupervised segmentation leads to a restoration procedure that is completely data driven and really compatible with an automatic processing of massive amounts of 3D SPECT data. (slides)







| References |
- M. Mignotte, J. Meunier, J.-C. Tardif. Endocardial Boundary estimation and tracking in echocardiographic images using deformable templates and Markov Random Fields. Pattern Analysis and Applications, 4(4):256-271, November 2001. (postscript) (pdf) (abstract)
- M. Mignotte, J. Meunier. A multiscale optimization approach for the dynamic contour-based boundary detection issue. Computerized Medical Imaging and Graphics, 25(3):265-275, May 2001. (postscript) (pdf) (abstract)
- M. Mignotte, J. Meunier.Three-dimensional blind deconvolution of SPECT images. IEEE Transactions on Biomedical Engineering, 47(2):274-281, February 2000. (postscript) (pdf) (abstract)
Postdoctoral Report
- M. Mignotte. Bayesian inference and/or optimization strategies for some detection, traking, and deconvolution problems in medical images. INRIA postdoctoral Report, DIRO, Montreal University, Canada, Québec, September 1999. (postscript) (pdf)



