|
PLearn 0.1
|
|
PLearn 0.1
|
The first sentence should be a BRIEF DESCRIPTION of what the class does. More...
#include <ScoreLayerVariable.h>
Public Member Functions | |
| ScoreLayerVariable () | |
| Default constructor. | |
| PP< MoleculeTemplate > | getMoleculeTemplate (real molecule_id, real activity=-1) |
| Return the molecule template associated with the given (real number) molecule id and activity. | |
| PP< Molecule > | getMolecule (real molecule_id) |
| Return the molecule associated to the given (real number) molecule id. | |
| void | setMappingsSource (const VMat &source_vmat) |
| Obtain the mappings of the input variable from a VMatrix. | |
| void | setScalingCoefficient (int i, real coeff) |
| Modify scaling coefficient for i-th score to set it to 'coeff'. | |
| virtual void | recomputeSize (int &l, int &w) const |
| Recomputes the length l and width w that this variable should have, according to its parent variables. | |
| virtual void | fprop () |
| compute output given input | |
| virtual void | bprop () |
| virtual string | classname () const |
| virtual OptionList & | getOptionList () const |
| virtual OptionMap & | getOptionMap () const |
| virtual RemoteMethodMap & | getRemoteMethodMap () const |
| virtual ScoreLayerVariable * | deepCopy (CopiesMap &copies) const |
| virtual void | build () |
| Post-constructor. | |
| virtual void | makeDeepCopyFromShallowCopy (CopiesMap &copies) |
| Transforms a shallow copy into a deep copy. | |
Static Public Member Functions | |
| static string | _classname_ () |
| ScoreLayerVariable. | |
| static OptionList & | _getOptionList_ () |
| static RemoteMethodMap & | _getRemoteMethodMap_ () |
| static Object * | _new_instance_for_typemap_ () |
| static bool | _isa_ (const Object *o) |
| static void | _static_initialize_ () |
| static const PPath & | declaringFile () |
Public Attributes | |
| PP< RunICPVariable > | run_icp_var |
| The variable that will run the ICP alignments. | |
| PP< ChemicalICP > | icp_aligner_template |
| ### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //! | |
| int | n_active_templates |
| int | n_inactive_templates |
| bool | normalize_by_n_features |
| int32_t | seed_ |
| bool | simple_mixture |
| VMat | templates_source |
Static Public Attributes | |
| static StaticInitializer | _static_initializer_ |
Protected Member Functions | |
| int | getNActiveTemplates () |
| Return the actual number of active templates, i.e. | |
| int | getNInactiveTemplates () |
| Return the actual number of inactive templates, i.e. | |
Static Protected Member Functions | |
| static void | declareOptions (OptionList &ol) |
| Declares the class options. | |
Protected Attributes | |
| VMat | mappings_source |
| The VMatrix containing information about the input mappings. | |
| PP< PRandom > | random_gen |
| Random number generator. | |
| VarArray | outputs |
| The array of output variables, comprised of two parts: | |
| Var | final_output |
| The concatenation of the variables in 'outputs': this is what will be actually the value of this variable. | |
| hash_map< string, PP < MoleculeTemplate > > | molecule_templates |
| All molecule templates currently loaded, identified by their id string. | |
| hash_map< string, PP< Molecule > > | molecules |
| All molecules currently loaded, identified by their id string. | |
| int | n_active_in_source |
| Number of active molecules in the templates source. | |
| int | n_inactive_in_source |
| Number of inactive molecules in the templates source. | |
| VarArray | scaling_coeffs |
| The i-th element is a 1x1 Var that contains the normalization coefficient for the score of template number i. | |
Private Types | |
| typedef NaryVariable | inherited |
Private Member Functions | |
| void | build_ () |
| This does the actual building. | |
The first sentence should be a BRIEF DESCRIPTION of what the class does.
* ScoreLayerVariable * Place the rest of the class programmer documentation here. Doxygen supports Javadoc-style comments. See http://www.doxygen.org/manual.html
Definition at line 64 of file ScoreLayerVariable.h.
typedef NaryVariable PLearn::ScoreLayerVariable::inherited [private] |
Reimplemented from PLearn::NaryVariable.
Definition at line 66 of file ScoreLayerVariable.h.
| PLearn::ScoreLayerVariable::ScoreLayerVariable | ( | ) |
Default constructor.
Definition at line 70 of file ScoreLayerVariable.cc.
:
n_active_templates(-1),
n_inactive_templates(-1),
normalize_by_n_features(false),
seed_(-1),
simple_mixture(false),
random_gen(new PRandom())
{}
| string PLearn::ScoreLayerVariable::_classname_ | ( | ) | [static] |
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
| OptionList & PLearn::ScoreLayerVariable::_getOptionList_ | ( | ) | [static] |
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
| RemoteMethodMap & PLearn::ScoreLayerVariable::_getRemoteMethodMap_ | ( | ) | [static] |
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
| Object * PLearn::ScoreLayerVariable::_new_instance_for_typemap_ | ( | ) | [static] |
Reimplemented from PLearn::Object.
Definition at line 65 of file ScoreLayerVariable.cc.
| StaticInitializer ScoreLayerVariable::_static_initializer_ & PLearn::ScoreLayerVariable::_static_initialize_ | ( | ) | [static] |
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
| void PLearn::ScoreLayerVariable::bprop | ( | ) | [virtual] |
Implements PLearn::Variable.
Definition at line 147 of file ScoreLayerVariable.cc.
References final_output, getNActiveTemplates(), getNInactiveTemplates(), PLearn::Variable::gradientdata, i, PLearn::Variable::length(), n, PLASSERT, and simple_mixture.
{
// The gradient is back-propagated only on the score variables, since the
// other variables are inputs that do not need be updated.
int n = simple_mixture ? 1
: getNActiveTemplates() + getNInactiveTemplates();
PLASSERT( n <= length() );
real* copy_grad_ptr = final_output->gradientdata;
real* grad_ptr = gradientdata;
for (int i = 0; i < n; i++, copy_grad_ptr++, grad_ptr++)
*copy_grad_ptr += *grad_ptr;
}
| void PLearn::ScoreLayerVariable::build | ( | ) | [virtual] |
Post-constructor.
The normal implementation should call simply inherited::build(), then this class's build_(). This method should be callable again at later times, after modifying some option fields to change the "architecture" of the object.
Reimplemented from PLearn::NaryVariable.
Definition at line 163 of file ScoreLayerVariable.cc.
References PLearn::NaryVariable::build(), and build_().
{
inherited::build();
build_();
}
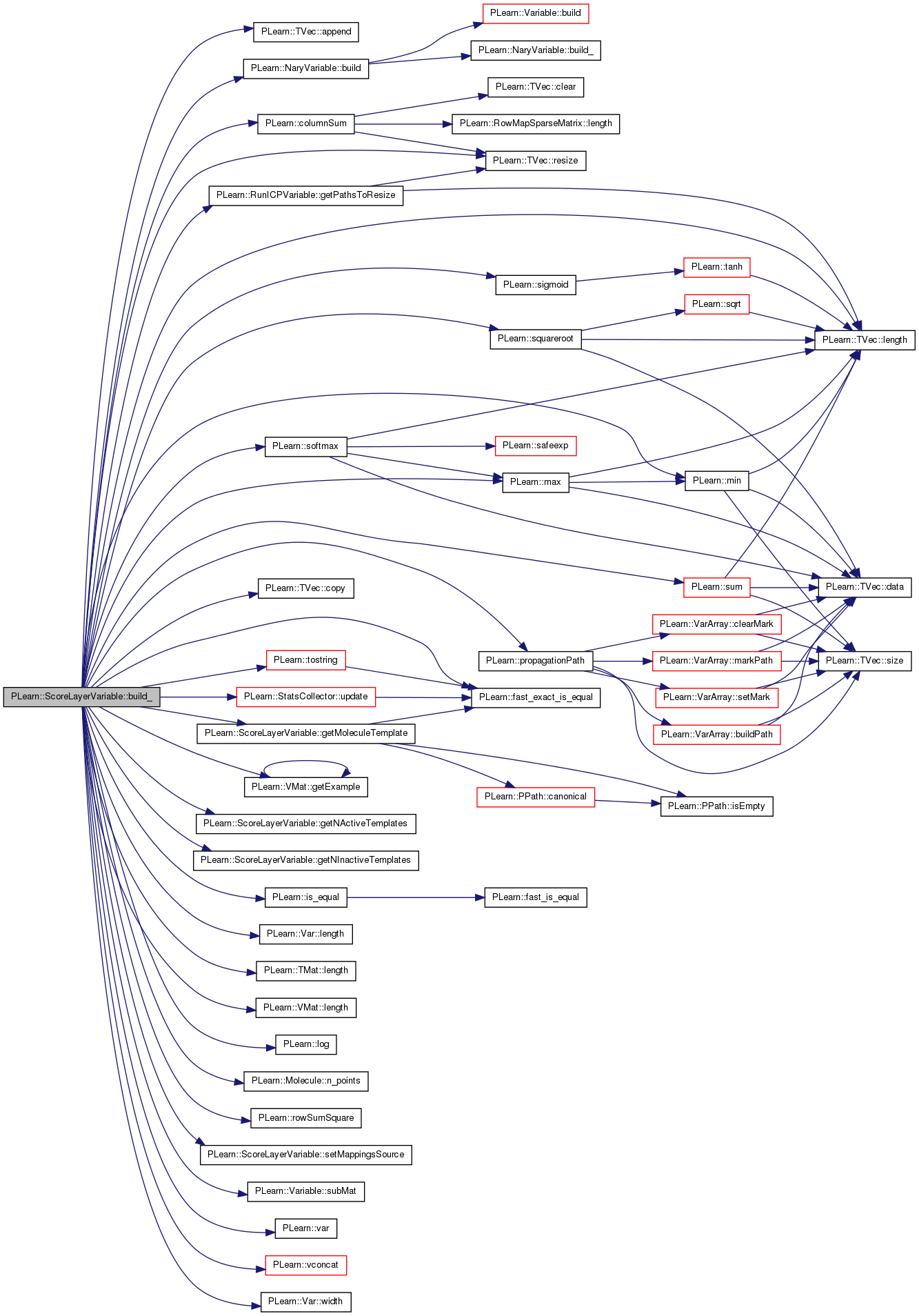
| void PLearn::ScoreLayerVariable::build_ | ( | ) | [private] |
This does the actual building.
Reimplemented from PLearn::NaryVariable.
Definition at line 172 of file ScoreLayerVariable.cc.
References PLearn::TVec< T >::append(), PLearn::NaryVariable::build(), PLearn::columnSum(), PLearn::TVec< T >::copy(), PLearn::fast_exact_is_equal(), PLearn::Molecule::feature_names, final_output, PLearn::VMat::getExample(), getMoleculeTemplate(), getNActiveTemplates(), getNInactiveTemplates(), PLearn::RunICPVariable::getPathsToResize(), i, icp_aligner_template, PLearn::is_equal(), j, PLearn::Var::length(), PLearn::TMat< T >::length(), PLearn::VMat::length(), PLearn::TVec< T >::length(), PLearn::log(), mappings_source, PLearn::max(), PLearn::min(), n, n_active_in_source, n_active_templates, n_inactive_in_source, n_inactive_templates, PLearn::Molecule::n_points(), outputs, PLASSERT, PLERROR, PLearn::propagationPath(), random_gen, PLearn::TVec< T >::resize(), PLearn::rowSumSquare(), run_icp_var, scaling_coeffs, seed_, setMappingsSource(), PLearn::sigmoid(), simple_mixture, PLearn::softmax(), PLearn::squareroot(), PLearn::Variable::subMat(), PLearn::sum(), templates_source, PLearn::tostring(), PLearn::StatsCollector::update(), PLearn::var(), PLearn::Variable::Var, PLearn::NaryVariable::varray, PLearn::vconcat(), and PLearn::Var::width().
Referenced by build().
{
if (seed_ != 0)
random_gen->manual_seed(seed_);
if (!templates_source)
return;
// Verify that we have been given the input variable, and resize the input
// array that is going to be constructed.
if (varray.length() < 1)
return;
varray.resize(1);
// Obtain mappings from the templates VMat in order to be able to load the
// molecule templates.
VMat mappings_source_backup = mappings_source;
setMappingsSource(templates_source);
PLASSERT( templates_source->targetsize() == 1 );
// Randomly select active and inactive templates.
TVec<int> list_of_active, list_of_inactive;
int n = templates_source->length();
Vec input, target;
real weight;
map<string, StatsCollector> chemical_stats;
for (int i = 0; i < n; i++) {
templates_source->getExample(i, input, target, weight);
PLASSERT( fast_exact_is_equal(target[0], 0) ||
fast_exact_is_equal(target[0], 1) );
if (fast_exact_is_equal(target[0], 0))
list_of_inactive.append(i);
else
list_of_active.append(i);
// Compute statistics on chemical features.
PP<MoleculeTemplate> mol_template =
getMoleculeTemplate(input[0], target[0]);
Mat& features = mol_template->features;
for (int j = 0; j < mol_template->feature_names.length(); j++) {
string feature_name = mol_template->feature_names[j];
StatsCollector& stats_col = chemical_stats[feature_name];
for (int k = 0; k < features.length(); k++)
stats_col.update(features(k, j));
}
}
n_active_in_source = list_of_active.length();
n_inactive_in_source = list_of_inactive.length();
if (n_active_templates != -1)
random_gen->shuffleElements(list_of_active);
if (n_inactive_templates != -1)
random_gen->shuffleElements(list_of_inactive);
PLASSERT( list_of_active.length() >= getNActiveTemplates() );
PLASSERT( list_of_inactive.length() >= getNInactiveTemplates() );
list_of_active.resize(getNActiveTemplates());
list_of_inactive.resize(getNInactiveTemplates());
// We do a copy of the list of active molecules instead of just appending
// the inactive ones because this list may be reused later (in the case of
// the simple mixture for instance).
TVec<int> templates = list_of_active.copy();
templates.append(list_of_inactive);
// Create the Var that will run all ICPs.
run_icp_var = new RunICPVariable(varray[0]);
run_icp_var->setScoreLayer(this);
// This VarArray will list additional parameters that must be optimized.
VarArray optimized_params;
// Create the corresponding score variables.
outputs.resize(0);
scaling_coeffs.resize(0);
int index_in_run_icp_var = 0; // Current index.
TVec<int> indices_of_active; // List of active templates.
for (int i = 0; i < templates.length(); i++) {
templates_source->getExample(templates[i], input, target, weight);
if (is_equal(target[0], 1))
indices_of_active.append(i);
PP<MoleculeTemplate> mol_template =
getMoleculeTemplate(input[0], target[0]);
Var molecule_coordinates(mol_template->n_points(), 3);
// Create the ICP aligner that will be used for this template.
CopiesMap copies;
PP<ChemicalICP> icp_aligner = icp_aligner_template->deepCopy(copies);
icp_aligner->mol_template = mol_template;
icp_aligner->build();
// Set the current molecule of the ICP aligner to be the same template:
// this will properly resize some important Vars.
icp_aligner->setMolecule((MoleculeTemplate*) mol_template);
// Initialize standard deviations of chemical features from the global
// standard deviations.
for (int j = 0; j < mol_template->feature_names.length(); j++) {
string feature_name = mol_template->feature_names[j];
icp_aligner->all_template_feat_dev->matValue.column(j).fill(
chemical_stats[feature_name].stddev());
}
// BC TODO ADD pout des stats
// Declare this new template (with associated molecule coordinates) to
// the RunICPVariable.
run_icp_var->addTemplate(icp_aligner, (MoleculeTemplate*) mol_template,
molecule_coordinates);
// Declare the RunICPVariable as parent of the feature indices, in
// order to ensure these variables are used after ICP has been run.
// Similarly, the variable containing the indices of the matching
// neighbors has to have the RunICPVariable as parent.
PP<UnaryVariable> mol_feat_indices =
(UnaryVariable*) ((Variable*) icp_aligner->mol_feat_indices);
mol_feat_indices->setInput((RunICPVariable*) run_icp_var);
PP<UnaryVariable> template_feat_indices =
(UnaryVariable*) ((Variable*) icp_aligner->template_feat_indices);
template_feat_indices->setInput((RunICPVariable*) run_icp_var);
PP<UnaryVariable> matching_neighbors =
(UnaryVariable*) ((Variable*) icp_aligner->matching_neighbors);
matching_neighbors->setInput((RunICPVariable*) run_icp_var);
// Build graph of Variables.
// (1) Compute the distance in chemical features.
Var template_features = icp_aligner->used_template_features;
icp_aligner->all_template_features->setName(
"all_template_features_" + tostring(i));
optimized_params.append(icp_aligner->all_template_features);
Var molecule_features_all_points = icp_aligner->used_mol_features;
Var molecule_features =
new VarRowsVariable(molecule_features_all_points,
matching_neighbors);
Var diff_features = template_features - molecule_features;
Var template_features_stddev = icp_aligner->used_template_feat_dev;
icp_aligner->all_template_feat_dev->setName(
"all_template_feat_dev_" + tostring(i));
optimized_params.append(icp_aligner->all_template_feat_dev);
Var feature_distance_at_each_point =
rowSumSquare(diff_features / template_features_stddev);
feature_distance_at_each_point->setName(
"feature_distance_at_each_point_" + tostring(i));
Var total_feature_distance = columnSum(feature_distance_at_each_point);
total_feature_distance->setName("total_feature_distance_"+tostring(i));
// (2) Compute the associated weights for the geometric distance.
string wm = icp_aligner->weighting_method;
Var weights;
if (wm == "none") {
weights = Var(mol_template->n_points(), 1);
weights->value.fill(1);
} else if (wm == "features_sigmoid") {
Var shift = icp_aligner->weighting_params[0];
Var slope = icp_aligner->weighting_params[1];
// We add a small value to avoid zero distances, as the square root
// is not differentiable at zero.
Var regularized_distances = feature_distance_at_each_point + 1e-10;
Var l1_dist = squareroot(regularized_distances);
weights = - sigmoid(slope * (l1_dist - shift));
shift->setName("shift_" + tostring(i));
slope->setName("slope_" + tostring(i));
weights->setName("weights_" + tostring(i));
optimized_params.append(shift);
optimized_params.append(slope);
} else {
PLERROR("In ScoreLayerVariable::build_ - Unsupported value for "
"'weighting_method'");
}
// (3) Compute the distance in geometric coordinates.
Var template_coordinates =
PLearn::subMat((RunICPVariable*) run_icp_var,
index_in_run_icp_var, 0,
mol_template->n_points(), 3);
index_in_run_icp_var += mol_template->n_points();
Var diff_coordinates = template_coordinates - molecule_coordinates;
Var template_coordinates_stddev = icp_aligner->template_geom_dev;
template_coordinates_stddev->setName(
"template_coordinates_stddev_" + tostring(i));
optimized_params.append(template_coordinates_stddev);
Var distance_at_each_point =
rowSumSquare(diff_coordinates / template_coordinates_stddev);
distance_at_each_point->setName("distance_at_each_point_"+tostring(i));
Var weighted_total_geometric_distance =
columnSum(distance_at_each_point * weights);
weighted_total_geometric_distance->setName(
"weighted_total_geometric_distance_" + tostring(i));
// (4) Sum to obtain the final score.
Var total_cost =
-0.5 * (total_feature_distance +
weighted_total_geometric_distance)
- sum(log(template_coordinates_stddev))
- sum(log(template_features_stddev));
// Note that at build time, the scaling coefficient is always equal to
// one over the number of points, regardless of the value of option
// 'normalize_by_n_features': this is because the number of features
// depends on the molecule being aligned, thus the scaling coefficient
// will be modified at run time by the RunICPVariable.
Var scaling_var = var(real(1.0 / mol_template->n_points()));
scaling_coeffs.append(scaling_var);
total_cost = scaling_var * total_cost;
// (5) Compute path on which sizes will have to be updated after ICP.
VarArray path_inputs = icp_aligner->used_template_features &
icp_aligner->used_mol_features &
icp_aligner->used_template_feat_dev;
VarArray path_outputs = total_cost;
VarArray& path_to_resize = run_icp_var->getPathsToResize(i);
path_to_resize = propagationPath(path_inputs, path_outputs);
// (6) Add template coefficient in the case of the simple mixture mode.
if (simple_mixture) {
Var coeff = var(real(1.0 / templates.length()));
coeff->min_value = 1e-10;
coeff->max_value = 1;
total_cost += log(coeff);
optimized_params.append(coeff);
}
// (7) Build output array.
outputs.append(total_cost);
}
// Append the additional input features if they are present.
if (templates_source->inputsize() > 1) {
if (simple_mixture)
PLERROR("In ScoreLayerVariable::build_ - The simple mixture model "
"is not meant to be used with extra input information");
Var input_var = varray[0];
PLASSERT( input_var->width() == 1 );
Var input_minus_molecule_id =
PLearn::subMat(input_var, 1, 0, input_var->length() - 1, 1);
outputs.append(input_minus_molecule_id);
}
// Concatenate all outputs in a single Variable.
final_output = vconcat(outputs);
// Build softmax output for activity probability in the case of the simple
// mixture model.
if (simple_mixture) {
Var softmax_output = softmax(final_output);
PLASSERT( list_of_active.length() > 0 );
PLASSERT( softmax_output->width() == 1 );
// Select only active templates (we assume - and verify - here that
// they they are the first ones).
if (min(list_of_active) != 0 ||
max(list_of_active) != list_of_active.length() - 1)
{
PLERROR("In ScoreLayerVariable::build_ - The active templates must"
" be first");
}
final_output = PLearn::subMat(softmax_output, 0, 0,
list_of_active.length(), 1);
// Then we just sum over the resulting posteriors.
final_output = sum(final_output);
}
// The final 'varray' will contain, in this order:
// - the input variable to this layer (already here)
// - the parameters (means, standard deviations, ...)
// - the final output
// The final output is not a parameter that will be updated during
// initialization, but it needs to be in this Variable's parents so that
// back-propagation is correctly performed.
varray.append(optimized_params);
varray.append(final_output);
// We have changed a parent's option, we should re-build.
inherited::build();
// Restore the mappings source that was given prior to calling build().
if (mappings_source_backup)
setMappingsSource(mappings_source_backup);
}

| string PLearn::ScoreLayerVariable::classname | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 65 of file ScoreLayerVariable.cc.
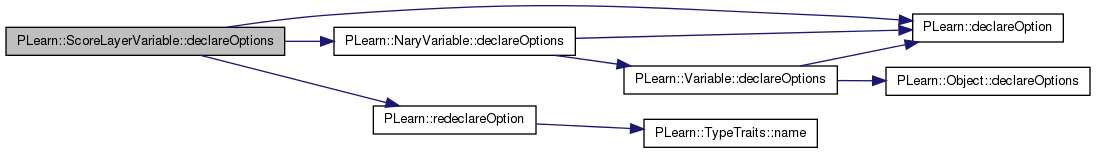
| void PLearn::ScoreLayerVariable::declareOptions | ( | OptionList & | ol | ) | [static, protected] |
Declares the class options.
Reimplemented from PLearn::NaryVariable.
Definition at line 82 of file ScoreLayerVariable.cc.
References PLearn::OptionBase::buildoption, PLearn::declareOption(), PLearn::NaryVariable::declareOptions(), icp_aligner_template, PLearn::OptionBase::learntoption, n_active_templates, n_inactive_templates, normalize_by_n_features, PLearn::redeclareOption(), seed_, simple_mixture, templates_source, PLearn::Variable::varname, and PLearn::NaryVariable::varray.
{
declareOption(ol, "icp_aligner_template",
&ScoreLayerVariable::icp_aligner_template,
OptionBase::buildoption,
"The model of ICP aligner we want to use (will be replicated for\n"
"each underlying score variable).");
declareOption(ol, "n_active_templates",
&ScoreLayerVariable::n_active_templates,
OptionBase::buildoption,
"Number of randomly chosen templates of active molecules (-1 means\n"
"we take all of them and they are not shuffled).");
declareOption(ol, "n_inactive_templates",
&ScoreLayerVariable::n_inactive_templates,
OptionBase::buildoption,
"Number of randomly chosen templates of inactive molecules (-1 means\n"
"we take all of them and they are not shuffled).");
declareOption(ol, "normalize_by_n_features",
&ScoreLayerVariable::normalize_by_n_features,
OptionBase::buildoption,
"If true, the score will be normalized by the number of features,\n"
"i.e. scaled by one over (3 + number of common chemical features).");
declareOption(ol, "seed", &ScoreLayerVariable::seed_,
OptionBase::buildoption,
"Seed of the random number generator (similar to a PLearner's seed).");
declareOption(ol, "templates_source",
&ScoreLayerVariable::templates_source,
OptionBase::learntoption, // To simplify help.
"The VMat templates are obtained from. This VMat's first column must\n"
"be the name of a molecule, there may be other input features, and\n"
"there must be a binary target indicating whether a molecule is\n"
"active or inactive. This VMat is also used to initialize standard\n"
"deviations of chemical features.");
// It is a learnt option as usually it will be set by the
// SurfaceTemplateLearner above.
declareOption(ol, "simple_mixture", &ScoreLayerVariable::simple_mixture,
OptionBase::learntoption,
"If true, then instead of being a layer of scores, it will be a\n"
"single-unit layer where scores have been processed to compute an\n"
"activity probability, by adding a log coefficient to each template,\n"
"performing a softmax on the resulting scores and summing over the\n"
"weights obtained for active templates.");
// Now call the parent class' declareOptions
inherited::declareOptions(ol);
// Redeclare some options as learnt options to make help simpler.
redeclareOption(ol, "varray", &ScoreLayerVariable::varray,
OptionBase::learntoption,
"Now a learnt option to simplify help.");
redeclareOption(ol, "varname", &ScoreLayerVariable::varname,
OptionBase::learntoption,
"Now a learnt option to simplify help.");
}
| static const PPath& PLearn::ScoreLayerVariable::declaringFile | ( | ) | [inline, static] |
| ScoreLayerVariable * PLearn::ScoreLayerVariable::deepCopy | ( | CopiesMap & | copies | ) | const [virtual] |
Reimplemented from PLearn::NaryVariable.
Definition at line 65 of file ScoreLayerVariable.cc.
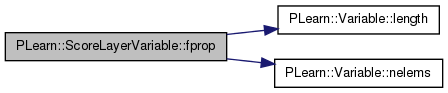
| void PLearn::ScoreLayerVariable::fprop | ( | ) | [virtual] |
compute output given input
Implements PLearn::Variable.
Definition at line 438 of file ScoreLayerVariable.cc.
References final_output, i, PLearn::Variable::length(), n, PLearn::Variable::nelems(), PLASSERT, and PLearn::Variable::valuedata.
{
// Just need to copy the data from 'final_output'.
int n = nelems();
PLASSERT( n == length() );
real* copy_ptr = final_output->valuedata;
real* ptr = valuedata;
for (int i = 0; i < n; i++, ptr++, copy_ptr++)
*ptr = *copy_ptr;
}
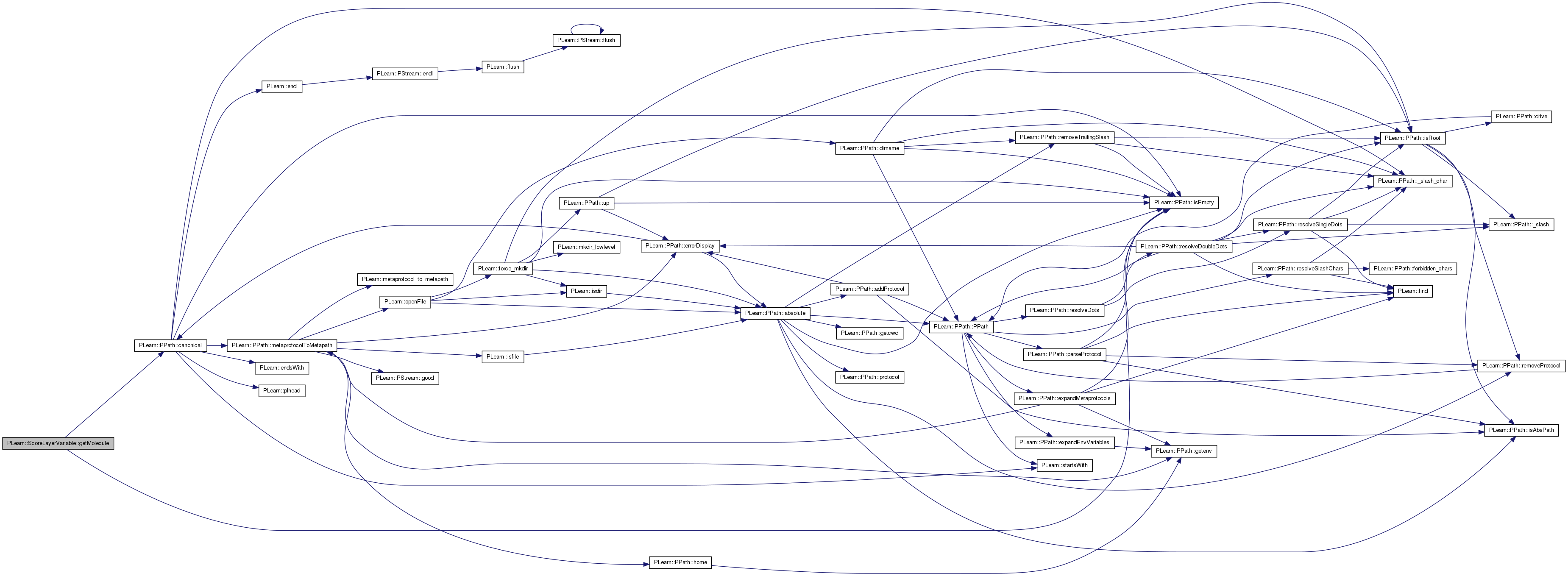
Return the molecule associated to the given (real number) molecule id.
Note that this method is not called from this class: it may not be the cleanest way to do it, but we need to cache the molecules somewhere to ensure they do not need to be systematically reloaded.
Definition at line 452 of file ScoreLayerVariable.cc.
References PLearn::PPath::canonical(), PLearn::PPath::isEmpty(), mappings_source, molecules, PLASSERT, and PLERROR.
Referenced by PLearn::RunICPVariable::fprop().
{
PLASSERT( mappings_source );
PPath molecule_path = mappings_source->getValString(0, molecule_id);
if (molecule_path.isEmpty())
PLERROR("In ScoreLayerVariable::getMolecule - Could not find "
"associated mapping");
string canonic_path = molecule_path.canonical();
// Load the molecule if necessary.
Molecule* molecule = 0;
if (molecules.find(canonic_path) == molecules.end()) {
molecule =
new Molecule(molecule_path);
molecules[canonic_path] = molecule;
} else
molecule = molecules[canonic_path];
PLASSERT( molecule );
return molecule;
}

| PP< MoleculeTemplate > PLearn::ScoreLayerVariable::getMoleculeTemplate | ( | real | molecule_id, |
| real | activity = -1 |
||
| ) |
Return the molecule template associated with the given (real number) molecule id and activity.
Definition at line 475 of file ScoreLayerVariable.cc.
References PLearn::PPath::canonical(), PLearn::fast_exact_is_equal(), PLearn::PPath::isEmpty(), mappings_source, molecule_templates, PLASSERT, and PLERROR.
Referenced by build_().
{
PLASSERT( fast_exact_is_equal(activity, 0) ||
fast_exact_is_equal(activity, 1) ||
fast_exact_is_equal(activity, -1) );
PLASSERT( mappings_source );
PPath molecule_path = mappings_source->getValString(0, molecule_id);
if (molecule_path.isEmpty())
PLERROR("In ScoreLayerVariable::getMolecule - Could not find "
"associated mapping");
string canonic_path = molecule_path.canonical();
// Load the molecule if necessary.
MoleculeTemplate* molecule = 0;
if (molecule_templates.find(canonic_path) == molecule_templates.end()) {
molecule =
new MoleculeTemplate(molecule_path, int(activity));
molecule_templates[canonic_path] = molecule;
} else
molecule = molecule_templates[canonic_path];
PLASSERT( molecule );
return molecule;
}

| int PLearn::ScoreLayerVariable::getNActiveTemplates | ( | ) | [protected] |
Return the actual number of active templates, i.e.
n_active_templates if it is non-negative, and the total number of active molecules in the templates source otherwise.
Definition at line 502 of file ScoreLayerVariable.cc.
References n_active_in_source, and n_active_templates.
Referenced by bprop(), and build_().
{
if (n_active_templates >= 0)
return n_active_templates;
else
return n_active_in_source;
}

| int PLearn::ScoreLayerVariable::getNInactiveTemplates | ( | ) | [protected] |
Return the actual number of inactive templates, i.e.
n_active_templates if it is non-negative, and the total number of inactive molecules in the templates source otherwise.
Definition at line 513 of file ScoreLayerVariable.cc.
References n_inactive_in_source, and n_inactive_templates.
Referenced by bprop(), and build_().
{
if (n_inactive_templates >= 0)
return n_inactive_templates;
else
return n_inactive_in_source;
}

| OptionList & PLearn::ScoreLayerVariable::getOptionList | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 65 of file ScoreLayerVariable.cc.
| OptionMap & PLearn::ScoreLayerVariable::getOptionMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 65 of file ScoreLayerVariable.cc.
| RemoteMethodMap & PLearn::ScoreLayerVariable::getRemoteMethodMap | ( | ) | const [virtual] |
Reimplemented from PLearn::Object.
Definition at line 65 of file ScoreLayerVariable.cc.
| void PLearn::ScoreLayerVariable::makeDeepCopyFromShallowCopy | ( | CopiesMap & | copies | ) | [virtual] |
Transforms a shallow copy into a deep copy.
Reimplemented from PLearn::NaryVariable.
Definition at line 524 of file ScoreLayerVariable.cc.
References PLearn::deepCopyField(), final_output, icp_aligner_template, PLearn::NaryVariable::makeDeepCopyFromShallowCopy(), mappings_source, molecule_templates, molecules, outputs, random_gen, run_icp_var, scaling_coeffs, templates_source, and PLearn::varDeepCopyField().
{
inherited::makeDeepCopyFromShallowCopy(copies);
deepCopyField(run_icp_var, copies);
deepCopyField(icp_aligner_template, copies);
deepCopyField(templates_source, copies);
deepCopyField(mappings_source, copies);
deepCopyField(random_gen, copies);
deepCopyField(outputs, copies);
varDeepCopyField(final_output, copies);
deepCopyField(molecule_templates, copies);
deepCopyField(molecules, copies);
deepCopyField(scaling_coeffs, copies);
}
Recomputes the length l and width w that this variable should have, according to its parent variables.
This is used for ex. by sizeprop() The default version stupidly returns the current dimensions, so make sure to overload it in subclasses if this is not appropriate.
Reimplemented from PLearn::Variable.
Definition at line 543 of file ScoreLayerVariable.cc.
References final_output, PLearn::Var::length(), and PLearn::Var::width().
{
if (final_output) {
l = final_output->length();
w = final_output->width();
} else {
l = w = 0;
}
}

| void PLearn::ScoreLayerVariable::setMappingsSource | ( | const VMat & | source_vmat | ) |
Obtain the mappings of the input variable from a VMatrix.
Definition at line 556 of file ScoreLayerVariable.cc.
References mappings_source.
Referenced by build_().
{
mappings_source = source_vmat;
}

Modify scaling coefficient for i-th score to set it to 'coeff'.
Definition at line 564 of file ScoreLayerVariable.cc.
References i, PLearn::TVec< T >::length(), PLearn::Variable::nelems(), PLASSERT, and scaling_coeffs.
Referenced by PLearn::RunICPVariable::fprop().
{
PLASSERT( i << scaling_coeffs.length() );
PLASSERT( scaling_coeffs[i]->nelems() == 1 );
scaling_coeffs[i]->value[0] = coeff;
}


Reimplemented from PLearn::NaryVariable.
Definition at line 123 of file ScoreLayerVariable.h.
Var PLearn::ScoreLayerVariable::final_output [protected] |
The concatenation of the variables in 'outputs': this is what will be actually the value of this variable.
Definition at line 146 of file ScoreLayerVariable.h.
Referenced by bprop(), build_(), fprop(), makeDeepCopyFromShallowCopy(), and recomputeSize().
### declare public option fields (such as build options) here Start your comments with Doxygen-compatible comments such as //!
Definition at line 78 of file ScoreLayerVariable.h.
Referenced by build_(), declareOptions(), and makeDeepCopyFromShallowCopy().
VMat PLearn::ScoreLayerVariable::mappings_source [protected] |
The VMatrix containing information about the input mappings.
Definition at line 134 of file ScoreLayerVariable.h.
Referenced by build_(), getMolecule(), getMoleculeTemplate(), makeDeepCopyFromShallowCopy(), and setMappingsSource().
hash_map< string, PP<MoleculeTemplate> > PLearn::ScoreLayerVariable::molecule_templates [protected] |
All molecule templates currently loaded, identified by their id string.
Definition at line 149 of file ScoreLayerVariable.h.
Referenced by getMoleculeTemplate(), and makeDeepCopyFromShallowCopy().
hash_map< string, PP<Molecule> > PLearn::ScoreLayerVariable::molecules [protected] |
All molecules currently loaded, identified by their id string.
Definition at line 152 of file ScoreLayerVariable.h.
Referenced by getMolecule(), and makeDeepCopyFromShallowCopy().
int PLearn::ScoreLayerVariable::n_active_in_source [protected] |
Number of active molecules in the templates source.
Definition at line 155 of file ScoreLayerVariable.h.
Referenced by build_(), and getNActiveTemplates().
Definition at line 79 of file ScoreLayerVariable.h.
Referenced by build_(), declareOptions(), and getNActiveTemplates().
int PLearn::ScoreLayerVariable::n_inactive_in_source [protected] |
Number of inactive molecules in the templates source.
Definition at line 158 of file ScoreLayerVariable.h.
Referenced by build_(), and getNInactiveTemplates().
Definition at line 80 of file ScoreLayerVariable.h.
Referenced by build_(), declareOptions(), and getNInactiveTemplates().
Definition at line 81 of file ScoreLayerVariable.h.
Referenced by declareOptions(), and PLearn::RunICPVariable::fprop().
VarArray PLearn::ScoreLayerVariable::outputs [protected] |
The array of output variables, comprised of two parts:
Definition at line 142 of file ScoreLayerVariable.h.
Referenced by build_(), and makeDeepCopyFromShallowCopy().
PP<PRandom> PLearn::ScoreLayerVariable::random_gen [protected] |
Random number generator.
Definition at line 137 of file ScoreLayerVariable.h.
Referenced by build_(), and makeDeepCopyFromShallowCopy().
The variable that will run the ICP alignments.
Definition at line 71 of file ScoreLayerVariable.h.
Referenced by build_(), and makeDeepCopyFromShallowCopy().
VarArray PLearn::ScoreLayerVariable::scaling_coeffs [protected] |
The i-th element is a 1x1 Var that contains the normalization coefficient for the score of template number i.
This normalization coefficient is equal to one over the number of points in the template, optionally also multiplied by one over the number of features common with the current aligned molecules (i.e. 3 + the number of common chemical features); this optional additional normalization depending on the 'normalize_by_n_features' option.
Definition at line 167 of file ScoreLayerVariable.h.
Referenced by build_(), makeDeepCopyFromShallowCopy(), and setScalingCoefficient().
Definition at line 82 of file ScoreLayerVariable.h.
Referenced by build_(), and declareOptions().
Definition at line 83 of file ScoreLayerVariable.h.
Referenced by bprop(), build_(), and declareOptions().
Definition at line 84 of file ScoreLayerVariable.h.
Referenced by build_(), declareOptions(), and makeDeepCopyFromShallowCopy().
 1.7.4
1.7.4